| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,590,339 – 2,590,431 |
| Length | 92 |
| Max. P | 0.617455 |

| Location | 2,590,339 – 2,590,431 |
|---|---|
| Length | 92 |
| Sequences | 14 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.22 |
| Shannon entropy | 0.63302 |
| G+C content | 0.54575 |
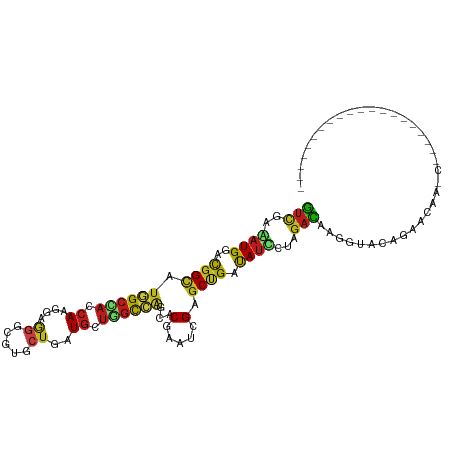
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.46 |
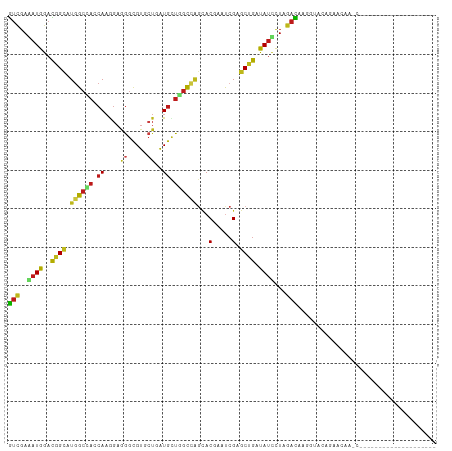
| Combinations/Pair | 1.68 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
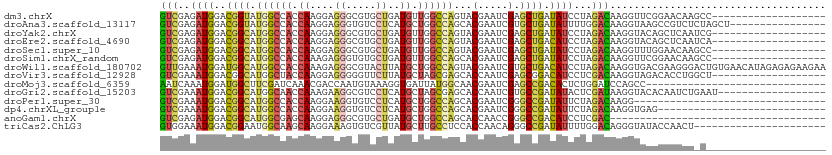
>dm3.chrX 2590339 92 - 22422827 GUCGAGAUGGACGGUAUGGCCACCAAGGAGGGCGUGCUGAUGUUGGCCAGUACGAAUCGAGCUGAUAUCCUAGACAAGGUUCGGAACAAGCC------------------- (((((.((...(((((((.((.(....).)).))))))))).)))))..((.((((((...(((......)))....))))))..)).....------------------- ( -26.00, z-score = 0.06, R) >droAna3.scaffold_13117 2523128 95 + 5790199 GUCGAGAUGGACGGUAUGGCCACCAAGGAGGGUGUCCUCAUGCUGGCCAGCACGAAUCGUGCUGAUAUUUUGGACAAGGUAAGCCGUCUCUAGCU---------------- ...(((((((.((((((((.((((......))))...))))))))..(((((((...))))))).((((((....))))))..))))))).....---------------- ( -35.40, z-score = -1.68, R) >droYak2.chrX 16010657 92 + 21770863 GUCGAGAUGGACGGCAUGGCCACCAAGGAGGGCGUGCUGAUGUUGGCCAGUACGAAUCGAGCUGAUAUCCUAGACAAGGUACAGCUCAAUCG------------------- (((((.((...(((((((.((.(....).)).))))))))).))))).....(((...((((((.((.(((.....)))))))))))..)))------------------- ( -32.20, z-score = -1.63, R) >droEre2.scaffold_4690 16545 92 - 18748788 GUCGAGAUGGACGGCAUGGCCACCAAGGAGGGCGUGCUGAUGUUGGCCAGUACGAAUCGAGCUGACAUCCUAGACAAGGUACAGCUCAAUCA------------------- (((((.((...(((((((.((.(....).)).))))))))).)))))...........((((((....(((.....)))..)))))).....------------------- ( -31.50, z-score = -1.22, R) >droSec1.super_10 2352035 92 - 3036183 GUCGAGAUGGACGGCAUGGCCACCAAGGAGGGCGUGCUGAUGUUGGCCAGUACGAAUCGAGCUGAUAUCCUAGACAAGGUUUGGAACAAGCC------------------- (((.....((((((((((.((.(....).)).))))))).(((..((..(.......)..))..))))))..)))..((((((...))))))------------------- ( -30.50, z-score = -1.40, R) >droSim1.chrX_random 1367358 92 - 5698898 GUCGAGAUGGACGGCAUGGCCACCAAAGAGGGUGUGCUGAUGUUGGCCAGCACGAAUCGAGCUGAUAUCCUAGACAAGGUUCGGAACAAGCC------------------- .(((.(.(((.(((((((((((((......)))).))).)))))).))).).)))...(..((((...(((.....))).))))..).....------------------- ( -28.30, z-score = -0.65, R) >droWil1.scaffold_180702 3677367 111 + 4511350 GUUGAAAUGGAUGGCAUGGCCACCAAAGAGGGCGUACUUAUGCUGGCCAGUACGAAUCGUGCUGACAUCCUAGACAAGGUGACGAAGGGACUGUGAACAUAGAGAGAAGAA (((.....(((((....(((((((.....))((((....)))))))))((((((...))))))..)))))..)))..(....).......((((....))))......... ( -29.70, z-score = -1.12, R) >droVir3.scaffold_12928 7353682 92 + 7717345 GUCGAAAUGGACGGCAUGGCUACCAAGGAGGGGGUUCUUAUGCUAGCGAGCACCAAUCGAGCGGACAUCCUCGACAAGGUAGACACCUGGCU------------------- (((((.(((..((((((((..(((........)))..))))))...(((.......)))..))..)))..))))).((((....))))....------------------- ( -26.10, z-score = 0.17, R) >droMoj3.scaffold_6359 2366455 81 + 4525533 AAUCAAAUGGAUGGCUUCGAUCAAACGACCAAUGUAAAGGUGAUUAUGGCAACGAAUCGAGCCGACACUCUGGAUCCAGCC------------------------------ .......(((((....(((......)))(((..((...(((((((.((....))))))..)))...))..))))))))...------------------------------ ( -17.90, z-score = -0.30, R) >droGri2.scaffold_15203 5191970 93 + 11997470 GUCGAAAUGGACGGCAUGGCAACCAAAGAAGGCGUCCUCAUGCUAGCGAGCACCAAUCGUGCCGAUAUACUCGAUAAGGUACACAAUCUGAAU------------------ (((((.(((..((((((((........(..(((((....)))))..).........)))))))).)))..))))).((((.....))))....------------------ ( -20.23, z-score = 0.42, R) >droPer1.super_30 615439 79 - 958394 GUCGAAAUGGACGGCAUGGCCACCAAGGAAGGUGUCCUCAUGCUGGCCAGCACGAAUCGGGCCGAUAUUCUAGACAAGG-------------------------------- (((..((((..((((.((((((.((((((.....))))..)).))))))...((...)).)))).))))...)))....-------------------------------- ( -24.80, z-score = -0.66, R) >dp4.chrXL_group1e 11927623 83 + 12523060 GUCGAAAUGGACGGCAUGGCCACCAAGGAAGGUGUCCUCAUGCUGGCCAGCACGAAUCGGGCCGAUAUUCUAGACAAGGUGAG---------------------------- (((..((((..((((.((((((.((((((.....))))..)).))))))...((...)).)))).))))...)))........---------------------------- ( -24.80, z-score = -0.12, R) >anoGam1.chrX 12207954 75 + 22145176 GUCGAGAUGGACGGCAUGGCGAGCAAGGAGGGCGUGCUGAUGCUGGCCAGCACCAACCGGGCCGACAUCCUCGAC------------------------------------ (((((((((..((((.((((.((((.((........))..)))).))))...((....)))))).))).))))))------------------------------------ ( -32.80, z-score = -1.92, R) >triCas2.ChLG3 21181840 89 + 32080666 GUGGAAAUGGACGGAAUGGCAAGCAAGGAAAGUGUCGUUAUGCUUGCCUCCACCAACAGGGCCGAUAUUUUGGACAGGGUAUACCAACU---------------------- .(((...(((..(((..(((((((((.((.....)).)..))))))))))).)))((...(((((....)))).)...))...)))...---------------------- ( -24.70, z-score = -0.43, R) >consensus GUCGAAAUGGACGGCAUGGCCACCAAGGAGGGCGUGCUGAUGCUGGCCAGCACGAAUCGAGCUGAUAUCCUAGACAAGGUACAGAACAA_C____________________ (((..((((..((((.((((((.((....((.....))..)).))))))...(.....).)))).))))...))).................................... (-13.68 = -13.22 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:50 2011