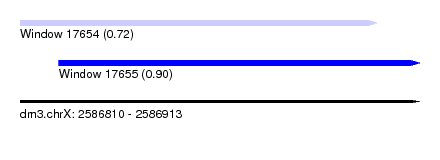
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,586,810 – 2,586,913 |
| Length | 103 |
| Max. P | 0.904961 |

| Location | 2,586,810 – 2,586,902 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 53.24 |
| Shannon entropy | 0.77196 |
| G+C content | 0.37004 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -6.64 |
| Energy contribution | -8.20 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.722000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2586810 92 + 22422827 AACACUUGUAGUAGUUGUUUUAAUCGAAGAUGGAUUGGAUUUAGCUG--CAAUUAGGUAGUUUAAUCAGUAAUUAGCUAGAGUGGCUAGUUAGA ((((((((..(((((((.(((((((.......)))))))..))))))--)...))))).))).......(((((((((.....))))))))).. ( -23.00, z-score = -2.12, R) >droSec1.super_10 2348339 92 + 3036183 CACACCUGGCCUAGUUGUUUCAAUCGGAUAUGGAUUGAGUGCAGCUAAAUAGUUAGUUAGUUUAAUCAGUAAUUAGCUAGAGUGGCAAGUAG-- .....((.(((((((((((((((((.......))))))).)))))))..((((((((((.(......).))))))))))....))).))...-- ( -26.30, z-score = -2.58, R) >droVir3.scaffold_12928 7349050 87 - 7717345 CAAGACUGGCCAGGUGUUAUCUUUUAUUCACAGUGGAAAAGCAAACACAUAUACAGAUCAC--AAUAUAUCUAUGUUUAAAAAUGUUAU----- ...(((((.....(((((..((((..(((.....)))))))..))))).....))).))..--..........................----- ( -10.60, z-score = 0.66, R) >droSim1.chrX_random 1364419 77 + 5698898 CACACCUGGCCUAGUUGUUUCAAUCGGAUAUGGAUUAGGUGCAGCUAGUUAGUUAGUU---------------UAGCUAGAGUGGCAAGUAG-- ..((((((((((((((((.((((((.......)))).)).)))))))).(((.....)---------------))))))).)))........-- ( -20.80, z-score = -1.09, R) >consensus CACACCUGGCCUAGUUGUUUCAAUCGGAUAUGGAUUAAAUGCAGCUA_AUAGUUAGUUAGU__AAUCAGUAAUUAGCUAGAGUGGCAAGUAG__ ..((((((...((((((((((((((.......))))))).)))))))..)))((((((((((........)))))))))).))).......... ( -6.64 = -8.20 + 1.56)

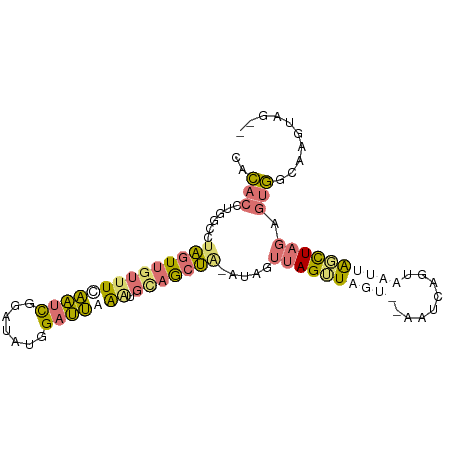
| Location | 2,586,820 – 2,586,913 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.40069 |
| G+C content | 0.38358 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2586820 93 + 22422827 GUAGUUGUUUUAAUCGAAGAUGGAUUGGAUUUAGCUG--CAAUUAGGUAGUUUAAUCAGUAAUUAGCUAGAGUGGCUAGUUAGACCGGACUUUGU ..............(((((.(((....((((.(((((--(......)))))).))))..(((((((((.....)))))))))..)))..))))). ( -23.80, z-score = -2.07, R) >droSec1.super_10 2348349 92 + 3036183 CUAGUUGUUUCAAUCGGAUAUGGAUUGAGUGCAGCUAAAUAGUUAGUUAGUUUAAUCAGUAAUUAGCUAGAGUGGCAAGU--AGUUCGACUUCG- ..............((((..(((((((..(((.(((...((((((((((.(......).)))))))))).))).)))..)--))))))..))))- ( -23.30, z-score = -2.27, R) >droSim1.chrX_random 1364429 77 + 5698898 CUAGUUGUUUCAAUCGGAUAUGGAUUAGGUGCAGCUAGUUAGUUAGUU---------------UAGCUAGAGUGGCAAGU--AGUUCGACUUCG- ..............((((..(((((((..(((..((((((((.....)---------------)))))))....)))..)--))))))..))))- ( -19.60, z-score = -1.66, R) >consensus CUAGUUGUUUCAAUCGGAUAUGGAUUGGGUGCAGCUA__UAGUUAGUUAGUUUAAUCAGUAAUUAGCUAGAGUGGCAAGU__AGUUCGACUUCG_ .((((((((((((((.......))))))).))))))).....((((((((((........))))))))))......................... (-13.14 = -13.73 + 0.59)

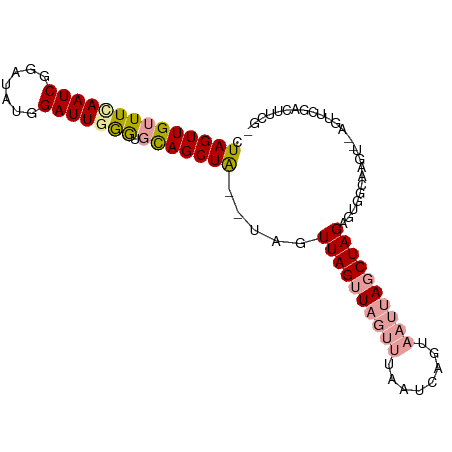
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:49 2011