| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,581,434 – 2,581,530 |
| Length | 96 |
| Max. P | 0.996279 |

| Location | 2,581,434 – 2,581,530 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.71 |
| Shannon entropy | 0.60150 |
| G+C content | 0.57983 |
| Mean single sequence MFE | -32.49 |
| Consensus MFE | -15.84 |
| Energy contribution | -18.05 |
| Covariance contribution | 2.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
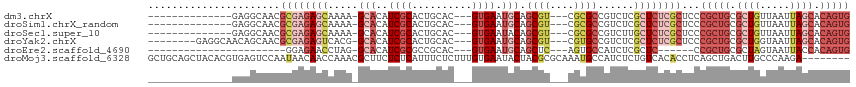
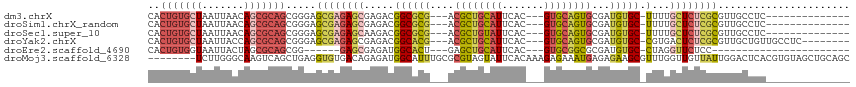
>dm3.chrX 2581434 96 + 22422827 --------------GAGGCAACGCGAGAGCAAAA-GCACAUCGCACUGCAC---GUGAAUGCAGCGU---CGCGCCGUCUCGCUCUCGCUCCCGCUGCGCUGUUAAUUAGCACAGUG --------------..(....)((((((((....-((.(..(((..((((.---.....))))))).---.).))......))))))))...(((((.((((.....)))).))))) ( -36.80, z-score = -0.96, R) >droSim1.chrX_random 1344717 96 + 5698898 --------------GAGGCAACGCGAGAGCAAAA-GCACAUCGCACUGCAC---GUGAAUGCAGCGU---CGCGCCGUCUCGCUCUCGCUCCCGCUGCGCUGUUAAUUAGCACAGUG --------------..(....)((((((((....-((.(..(((..((((.---.....))))))).---.).))......))))))))...(((((.((((.....)))).))))) ( -36.80, z-score = -0.96, R) >droSec1.super_10 2343011 96 + 3036183 --------------GAGGCAACGCGAGAGCAAAA-GCACAUCGCACUGCAC---GUGAAUACAGCGU---CGCGCCGUCUUGCUCUCGCUCCCGCUGCGCUGUUAAUUAGCACAGUG --------------..(....)((((((((((..-((.....))...((((---((.......))))---.))......))))))))))...(((((.((((.....)))).))))) ( -37.10, z-score = -1.66, R) >droYak2.chrX 16001670 102 - 21770863 --------GAGGCAACAGCAACGCGAGAGUCACG-GCACAUCGCACUGCAC---GUGAAUGCAGCGU---CGUGCCGUCUCGCUCUCGCUCCCGCUGCGCUGGUAAUUAGCACAGUG --------..(....)......((((((((.(((-((((..(((..((((.---.....))))))).---.)))))))...))))))))...(((((.((((.....)))).))))) ( -46.00, z-score = -2.57, R) >droEre2.scaffold_4690 7752 81 + 18748788 -----------------------GGAGAACCUAG-GCACAUCGCGCCGCAC---GUGAAUGCAGCUC---AGUGCCAUCUCGCUC------CCGCUGCGCUAGUAAUUACCACAGUG -----------------------((((......(-((((...(.((.(((.---.....))).)).)---.)))))......)))------)(((((.(..(....)...).))))) ( -22.00, z-score = 0.33, R) >droMoj3.scaffold_6328 471084 109 + 4453435 GCUGCAGCUACACGUGAGUCCAAUAACAACCAAACGCUUCUCUCAUUUCUCUUUGUGAAUACUACGCGCAAAUGCCAUCUCUGUCACACCUCAGCUGACUUGCCCAAGA-------- ((..(((((....(((((.......................))))).......(((((.......(.((....))).......)))))....)))))....))......-------- ( -16.24, z-score = 0.45, R) >consensus ______________GAGGCAACGCGAGAGCAAAA_GCACAUCGCACUGCAC___GUGAAUGCAGCGU___CGCGCCGUCUCGCUCUCGCUCCCGCUGCGCUGUUAAUUAGCACAGUG ......................((((((((.....((......(((........)))......(((....)))))......))))))))...(((((.((((.....)))).))))) (-15.84 = -18.05 + 2.21)
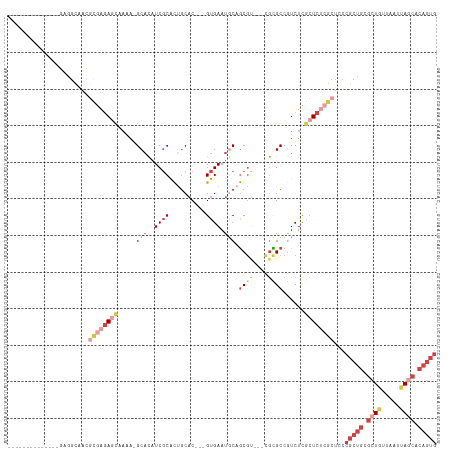
| Location | 2,581,434 – 2,581,530 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.71 |
| Shannon entropy | 0.60150 |
| G+C content | 0.57983 |
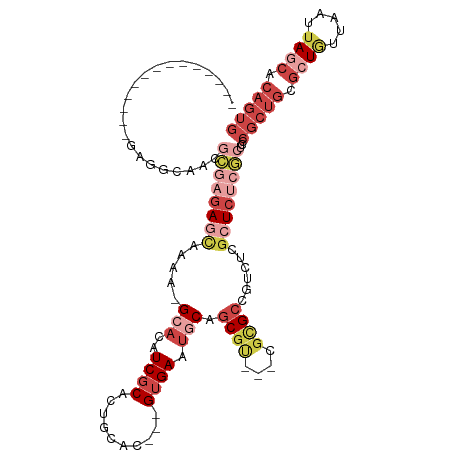
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -26.42 |
| Energy contribution | -28.17 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2581434 96 - 22422827 CACUGUGCUAAUUAACAGCGCAGCGGGAGCGAGAGCGAGACGGCGCG---ACGCUGCAUUCAC---GUGCAGUGCGAUGUGC-UUUUGCUCUCGCGUUGCCUC-------------- (.(((((((.......))))))).)((.(((((((((((..((((((---.((((((((....---))))))))...)))))-))))))))))))....))..-------------- ( -47.70, z-score = -2.96, R) >droSim1.chrX_random 1344717 96 - 5698898 CACUGUGCUAAUUAACAGCGCAGCGGGAGCGAGAGCGAGACGGCGCG---ACGCUGCAUUCAC---GUGCAGUGCGAUGUGC-UUUUGCUCUCGCGUUGCCUC-------------- (.(((((((.......))))))).)((.(((((((((((..((((((---.((((((((....---))))))))...)))))-))))))))))))....))..-------------- ( -47.70, z-score = -2.96, R) >droSec1.super_10 2343011 96 - 3036183 CACUGUGCUAAUUAACAGCGCAGCGGGAGCGAGAGCAAGACGGCGCG---ACGCUGUAUUCAC---GUGCAGUGCGAUGUGC-UUUUGCUCUCGCGUUGCCUC-------------- (.(((((((.......))))))).)((.(((((((((((..((((((---.((((((((....---))))))))...)))))-))))))))))))....))..-------------- ( -46.00, z-score = -3.05, R) >droYak2.chrX 16001670 102 + 21770863 CACUGUGCUAAUUACCAGCGCAGCGGGAGCGAGAGCGAGACGGCACG---ACGCUGCAUUCAC---GUGCAGUGCGAUGUGC-CGUGACUCUCGCGUUGCUGUUGCCUC-------- ...((((((.......))))))((((.(((((..(((((((((((((---.((((((((....---))))))))...)))))-))....)))))).))))).))))...-------- ( -49.10, z-score = -2.62, R) >droEre2.scaffold_4690 7752 81 - 18748788 CACUGUGGUAAUUACUAGCGCAGCGG------GAGCGAGAUGGCACU---GAGCUGCAUUCAC---GUGCGGCGCGAUGUGC-CUAGGUUCUCC----------------------- ..(((((.((.....)).))))).((------((((.....(((((.---..(((((((....---))))))).....))))-)...)))))).----------------------- ( -31.60, z-score = -1.08, R) >droMoj3.scaffold_6328 471084 109 - 4453435 --------UCUUGGGCAAGUCAGCUGAGGUGUGACAGAGAUGGCAUUUGCGCGUAGUAUUCACAAAGAGAAAUGAGAGAAGCGUUUGGUUGUUAUUGGACUCACGUGUAGCUGCAGC --------.(((.(((......))).))).............(((.((((((((....(((.((........)).)))..(.(((..((....))..))).))))))))).)))... ( -24.30, z-score = 1.68, R) >consensus CACUGUGCUAAUUAACAGCGCAGCGGGAGCGAGAGCGAGACGGCACG___ACGCUGCAUUCAC___GUGCAGUGCGAUGUGC_UUUGGCUCUCGCGUUGCCUC______________ (.(((((((.......))))))).)...((((((((......(((((....((((((((.......))))))))...))))).....))))))))...................... (-26.42 = -28.17 + 1.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:47 2011