| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,803,494 – 9,803,586 |
| Length | 92 |
| Max. P | 0.711306 |

| Location | 9,803,494 – 9,803,586 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Shannon entropy | 0.38095 |
| G+C content | 0.36654 |
| Mean single sequence MFE | -15.46 |
| Consensus MFE | -12.57 |
| Energy contribution | -12.39 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.01 |
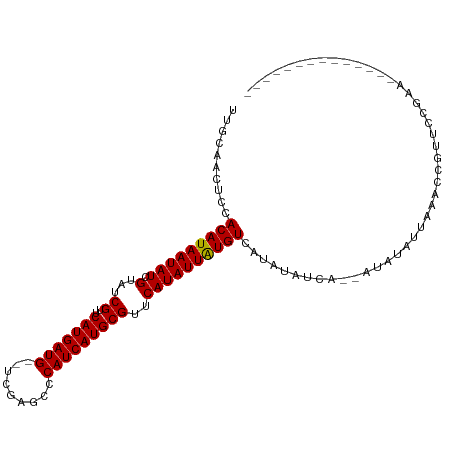
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
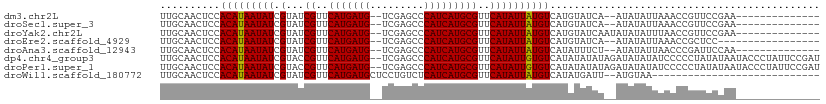
>dm3.chr2L 9803494 92 - 23011544 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUGUAUCA--AUAUAUUAAACCGUUCCGAA-------------- (((.(((...(((((((((.(...((..(((((((--.......)))))))))..)))))))))).(((((...--.)))))......))).))).-------------- ( -15.30, z-score = -0.79, R) >droSec1.super_3 5250249 92 - 7220098 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUGUAUCA--AUAUAUUAAACCGUUCCGAA-------------- (((.(((...(((((((((.(...((..(((((((--.......)))))))))..)))))))))).(((((...--.)))))......))).))).-------------- ( -15.30, z-score = -0.79, R) >droYak2.chr2L 12490017 94 - 22324452 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUGUAUCAAUAUAUAUUUAACCGUUCCGAA-------------- (((.(((...(((((((((.(...((..(((((((--.......)))))))))..)))))))))).((((((....))))))......))).))).-------------- ( -16.10, z-score = -0.97, R) >droEre2.scaffold_4929 10415972 89 - 26641161 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUGUAUCA--AUAUAUUAAACCGCUCC----------------- ..((......(((((((((.(...((..(((((((--.......)))))))))..)))))))))).(((((...--.)))))......))...----------------- ( -15.40, z-score = -1.42, R) >droAna3.scaffold_12943 1241600 92 - 5039921 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUAUUUCU--AUAUAUUAACCCGAUUCCAA-------------- ..........(((((((((.(...((..(((((((--.......)))))))))..)))))))))).........--....................-------------- ( -14.00, z-score = -1.39, R) >dp4.chr4_group3 11322777 108 + 11692001 UUGCAACUCCACAUAAUAUCGUACCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUGUGUCAUAUAUAUAGAUAUAUAUCCCCCUAUAUAAUACCCUAUUCCGAU ..........(((((((((.(...((..(((((((--.......)))))))))..))))))))))....(((((((............)))))))............... ( -16.90, z-score = -0.89, R) >droPer1.super_1 8438211 108 + 10282868 UUGCAACUCCACAUAAUAUCGUACCGUUCAUGAUG--UCGAGCCCAUCAUGCGUUCAUAUUGUGUCAUAUAUAUAGAUAUAUAUCCCCCUAUAUAAUACCCUAUUCCGAU ..........(((((((((.(...((..(((((((--.......)))))))))..))))))))))....(((((((............)))))))............... ( -16.90, z-score = -0.89, R) >droWil1.scaffold_180772 8450575 80 - 8906247 UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUGCUCCUGUCUCAUCAUGCGUUCAUAUUAUGUCAUAUGAUU--AUGUAA---------------------------- ..........(((((((((.(...(((..((((.((....)).))))...)))..))))))))))((((....)--)))...---------------------------- ( -13.80, z-score = -0.93, R) >consensus UUGCAACUCCACAUAAUAUCGUAUCGUUCAUGAUG__UCGAGCCCAUCAUGCGUUCAUAUUAUGUCAUAUAUCA__AUAUAUUAAACCGUUCCGAA______________ ..........(((((((((.(...((..(((((((.........)))))))))..))))))))))............................................. (-12.57 = -12.39 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:58 2011