
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,546,238 – 2,546,334 |
| Length | 96 |
| Max. P | 0.500000 |

| Location | 2,546,238 – 2,546,334 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Shannon entropy | 0.25301 |
| G+C content | 0.32836 |
| Mean single sequence MFE | -16.04 |
| Consensus MFE | -10.07 |
| Energy contribution | -10.46 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 2546238 96 + 22422827 AACUACUUGAGUGGAAUCUUGGUAAAGUUUAUUUACUAUACCCAGAAUAAUAUUGAUUUGGUUUUCUUGCUUUUCUACUCUAGUACUGCGUUCAUG- ...((((.(((((((((((.((((.(((......))).)))).))).............(((......))).)))))))).))))...........- ( -22.00, z-score = -2.83, R) >droSec1.super_10 2311904 74 + 3036183 AACUACUUGA-UGGAAUCUUGG-AAAGUUUAUUUACUA--CCCAGAAUAAUAUU-------------------UCUACUCGAGUACUGCGUUCAUAC ...(((((((-((((((((.((-..(((......))).--.))))).......)-------------------)))).)))))))............ ( -12.91, z-score = -1.39, R) >droSim1.chrX_random 1321775 74 + 5698898 AACUACUUGA-UGGAAUCUUGG-AAAGUUUAUUUACUA--CCCAGAAUAAUAUU-------------------UCUACUCAAGUACUGCGUUCAUAC ...(((((((-((((((((.((-..(((......))).--.))))).......)-------------------)))).)))))))............ ( -13.21, z-score = -1.71, R) >consensus AACUACUUGA_UGGAAUCUUGG_AAAGUUUAUUUACUA__CCCAGAAUAAUAUU___________________UCUACUCAAGUACUGCGUUCAUAC ...(((((((.((((.(((.(((..(((......)))..))).)))...........................)))).)))))))............ (-10.07 = -10.46 + 0.39)

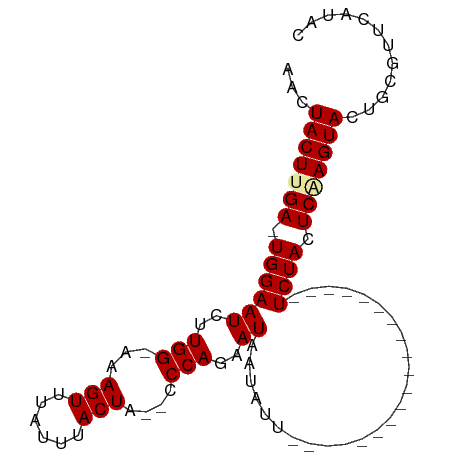

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:42 2011