| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,527,943 – 2,528,030 |
| Length | 87 |
| Max. P | 0.923076 |

| Location | 2,527,943 – 2,528,030 |
|---|---|
| Length | 87 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.49284 |
| G+C content | 0.47478 |
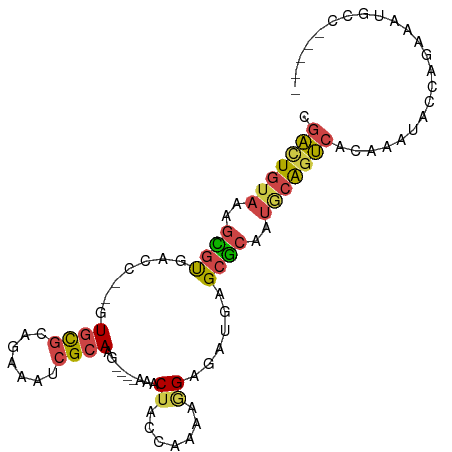
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -10.77 |
| Energy contribution | -10.83 |
| Covariance contribution | 0.06 |
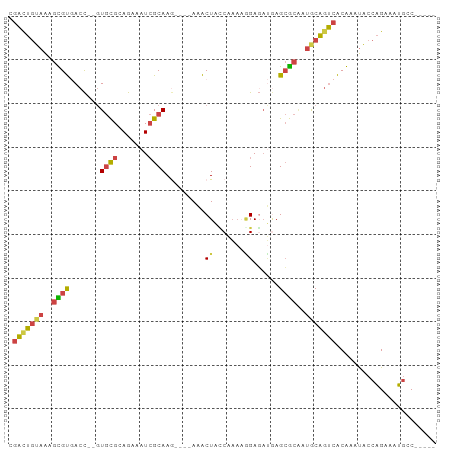
| Combinations/Pair | 1.53 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
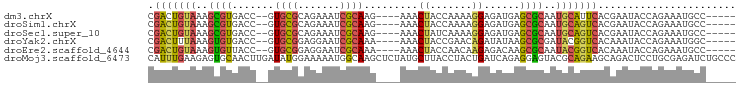
>dm3.chrX 2527943 87 - 22422827 CGACUGUAAAGCGUGACC--GUGCGCAGAAAUCGCAAG----AAACUACCAAAAGGAGAUGAGCGCAAUGCAUUCACGAAUACCAGAAAUGCC----- ...(((.....(((((..--.(((((.....((....)----)..((.((....))))....)))))......))))).....))).......----- ( -22.00, z-score = -2.32, R) >droSim1.chrX 1777472 87 - 17042790 CGACUGUAAAGCGUGACC--GUGCGCAGAAAUCGCAAG----AAACUACCAAAAGGAGAUGAGCGCAAUGCAGUCACGAAUACCAGAAAUGCC----- ...(((.....((((((.--.(((((.....((....)----)..((.((....))))....))))).....)))))).....))).......----- ( -25.10, z-score = -3.28, R) >droSec1.super_10 2293919 87 - 3036183 CGACUGUAAAGCGUGACC--GUGCGCAGAAAUCGCAAG----AAACUAUCAAAAGGAGAUGAGCGCAAUGCAGUCACGAAUACCAGAAAUGCC----- ...(((.....((((((.--.(((((.....((....)----)...((((.......)))).))))).....)))))).....))).......----- ( -24.10, z-score = -2.60, R) >droYak2.chrX 15945929 87 + 21770863 CGACUUUAAAGUGUGACC--GUGCGGAGGAAUCGCAAA----AAACUACCGAACAGAUAUAAGCGCGAUACGGUCACAAAUACCAGAAAUGGC----- ...........(((((((--((((((.....))))...----.......((..(........)..))..)))))))))....(((....))).----- ( -22.00, z-score = -2.28, R) >droEre2.scaffold_4644 2473827 87 - 2521924 CGACUGUAAAGUGUUACC--GUGCGGAGGAAUCGCAAA----AAACUACCAACAAGAGACAAGCGCAAUACGGUCACAAAUACCAGAAAUGCC----- .(((((((..(((((...--.(((((.....)))))..----...((.......)).....)))))..)))))))........((....))..----- ( -17.20, z-score = -0.90, R) >droMoj3.scaffold_6473 8526435 98 - 16943266 CAUUUGAAGAGUGCAACUUGAUAUGGAAAAAUGGCAAGCUCUAUGCUUACCUACUGAUCAGAGGAGUACGCAGAAGCAGACUCCUGCGAGAUCUGCCC ..........((((..((((((.(((......((.((((.....)))).))..))))))).))..))))(((((.((((....))))....))))).. ( -25.10, z-score = -0.80, R) >consensus CGACUGUAAAGCGUGACC__GUGCGCAGAAAUCGCAAG____AAACUACCAAAAGGAGAUGAGCGCAAUGCAGUCACAAAUACCAGAAAUGCC_____ .(((((((..((((.......((((.......)))).........((.......))......))))..)))))))....................... (-10.77 = -10.83 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:37 2011