
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,793,438 – 9,793,543 |
| Length | 105 |
| Max. P | 0.992941 |

| Location | 9,793,438 – 9,793,543 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.32421 |
| G+C content | 0.28104 |
| Mean single sequence MFE | -17.84 |
| Consensus MFE | -12.17 |
| Energy contribution | -12.81 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
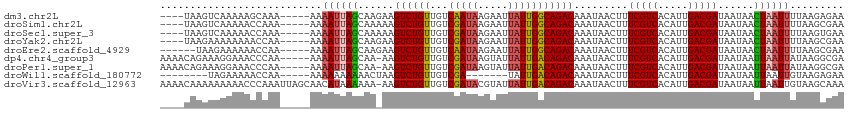
>dm3.chr2L 9793438 105 + 23011544 UUCUCUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUUGACAACAGACUUCUUGCUAAUUUU-----UUUGCUUUUUGACUUA---- .......(((((((((.....(((((.....))))).........(((((.(((((.....)))))....))))).....)))))))))-----................---- ( -18.60, z-score = -2.25, R) >droSim1.chr2L 9575326 105 + 22036055 UUCGCUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUCGACAACAGACUUUUUGCUAAUUUU-----UUUGGUUUUUGACUUA---- ...(((.(((((((((.....(((((.....)))))(((((..(((((.................)))))..)))))...)))))))))-----...)))..........---- ( -19.03, z-score = -1.59, R) >droSec1.super_3 5240463 105 + 7220098 UUCACUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUCGACAACAGACUUUUUGCUAAUUUU-----UUUGGUUUUUGACUUA---- ...(((.(((((((((.....(((((.....)))))(((((..(((((.................)))))..)))))...)))))))))-----...)))..........---- ( -18.73, z-score = -1.73, R) >droYak2.chr2L 12479808 105 + 22324452 UUCGCUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUCGACAACAGACUUCUUGCUAAUUUU-----UUGGUUUUUUUUCUUA---- ...(((.(((((((((.....(((((.....)))))(((((..(((((.................)))))..)))))...)))))))))-----..)))...........---- ( -19.03, z-score = -2.11, R) >droEre2.scaffold_4929 10405859 103 + 26641161 UUCGCUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUUGACAACAGACUUCUUGCUAAUUUU-----UUGGUUUUUUCUUA------ ...(((.(((((((((.....(((((.....))))).........(((((.(((((.....)))))....))))).....)))))))))-----..))).........------ ( -20.60, z-score = -2.76, R) >dp4.chr4_group3 11312683 108 - 11692001 UCGCCUUAUAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACUUAUCGACAACAGACUU-UUGCUAAUUUU-----UUGGGUUUCCUUUCUGUUUU ..((((...(((((.......(((((.....)))))......(((((.((((.(((.....))).)))))))))...-....)))))..-----..)))).............. ( -17.60, z-score = -1.26, R) >droPer1.super_1 8427746 108 - 10282868 UCGCCUUAUAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACUUAUCGACAACAGACUU-UUGCUAAUUUU-----UUGGGUUUCCUUUCUGUUUU ..((((...(((((.......(((((.....)))))......(((((.((((.(((.....))).)))))))))...-....)))))..-----..)))).............. ( -17.60, z-score = -1.26, R) >droWil1.scaffold_180772 8436644 94 + 8906247 UUCUCUUACAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUA-------UCGACAACAGACUUAGUUUUUUUUUU-----UUGGUUUUUCUA-------- ........(((..((..((((..(((...(((((....))))).(((.((((....-------..)))))))))).)))).....))..-----))).........-------- ( -13.90, z-score = -0.90, R) >droVir3.scaffold_12963 18387146 113 - 20206255 UUUGCUUACAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACGUAUCGACAACAGACUU-UUUUUUAUGUUGCUAAUUUGGGUUUUUUUUUGUUUU ...(((((.(((((.......(((((.....)))))......(((((.((((.(((.....))).)))))))))...-.............))))))))))............. ( -15.50, z-score = 0.40, R) >consensus UUCGCUUAAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUCGACAACAGACUU_UUGCUAAUUUU_____UUGGGUUUUUUUCUUA____ .........(((((((.....(((((.....)))))(((((..(((((.................)))))..)))))...)))))))........................... (-12.17 = -12.81 + 0.64)


| Location | 9,793,438 – 9,793,543 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.32421 |
| G+C content | 0.28104 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -16.14 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
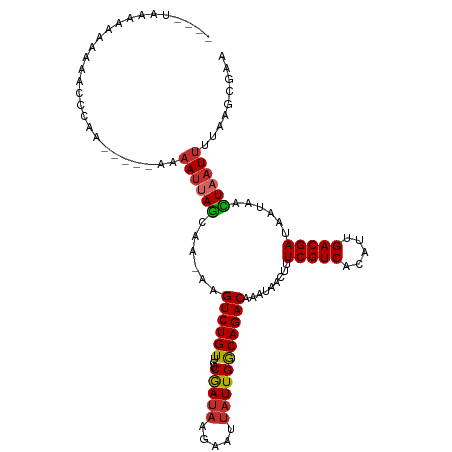
>dm3.chr2L 9793438 105 - 23011544 ----UAAGUCAAAAAGCAAA-----AAAAUUAGCAAGAAGUCUGUUGUCAAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGAGAA ----................-----((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))))....... ( -20.40, z-score = -2.78, R) >droSim1.chr2L 9575326 105 - 22036055 ----UAAGUCAAAAACCAAA-----AAAAUUAGCAAAAAGUCUGUUGUCGAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGCGAA ----................-----((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))))....... ( -20.10, z-score = -2.72, R) >droSec1.super_3 5240463 105 - 7220098 ----UAAGUCAAAAACCAAA-----AAAAUUAGCAAAAAGUCUGUUGUCGAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGUGAA ----............((..-----((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))))...)).. ( -20.50, z-score = -2.80, R) >droYak2.chr2L 12479808 105 - 22324452 ----UAAGAAAAAAAACCAA-----AAAAUUAGCAAGAAGUCUGUUGUCGAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGCGAA ----................-----((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))))....... ( -20.10, z-score = -2.78, R) >droEre2.scaffold_4929 10405859 103 - 26641161 ------UAAGAAAAAACCAA-----AAAAUUAGCAAGAAGUCUGUUGUCAAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGCGAA ------..............-----((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))))....... ( -20.40, z-score = -3.19, R) >dp4.chr4_group3 11312683 108 + 11692001 AAAACAGAAAGGAAACCCAA-----AAAAUUAGCAA-AAGUCUGUUGUCGAUAAGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUAUAAGGCGA ..........((....))..-----.......((..-..((((((((..(((....)))..)))))))).........(((((.....))))).................)).. ( -18.90, z-score = -2.03, R) >droPer1.super_1 8427746 108 + 10282868 AAAACAGAAAGGAAACCCAA-----AAAAUUAGCAA-AAGUCUGUUGUCGAUAAGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUAUAAGGCGA ..........((....))..-----.......((..-..((((((((..(((....)))..)))))))).........(((((.....))))).................)).. ( -18.90, z-score = -2.03, R) >droWil1.scaffold_180772 8436644 94 - 8906247 --------UAGAAAAACCAA-----AAAAAAAAAACUAAGUCUGUUGUCGA-------UAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUGUAAGAGAA --------............-----..............((((((((....-------...)))))))).........(((((.....)))))..................... ( -15.00, z-score = -1.84, R) >droVir3.scaffold_12963 18387146 113 + 20206255 AAAACAAAAAAAAACCCAAAUUAGCAACAUAAAAAA-AAGUCUGUUGUCGAUACGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUGUAAGCAAA .......................((...........-..((((((((..(((....)))..)))))))).........(((((.....)))))................))... ( -18.00, z-score = -2.35, R) >consensus ____UAAAAAAAAAACCCAA_____AAAAUUAGCAA_AAGUCUGUUGUCGAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUUAAGCGAA ...........................((((((......((((((...(((((.....))))))))))).........(((((.....)))))......))))))......... (-16.14 = -15.97 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:57 2011