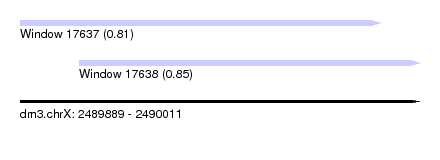
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,489,889 – 2,490,011 |
| Length | 122 |
| Max. P | 0.851129 |

| Location | 2,489,889 – 2,489,999 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Shannon entropy | 0.28233 |
| G+C content | 0.58149 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2489889 110 + 22422827 AGCCAGGAGCUGAUAGAGCAUC-----CGGUGCACCACAUA--UAUUGACCUGGGGG-UGCCUGGACCAAAACAACUUCCGUUCCUCUCGGCGGCAAUCGCUGAGCUUUCAGGCACAA .(((((((((.((.......((-----(((.(((((.((..--........))..))-))))))))...........)).))))))(((((((.....)))))))......))).... ( -37.97, z-score = -1.06, R) >droSim1.chrX 1766302 107 + 17042790 AGCCAGGAGCUGAUAGAGCAUC-----CGGAGCACCACACA--UAUUGACCUGGGG--UGCCUGGACCAAC-CAACUUCCGUUCCGCUCGGCGGCAAUCGCUGAGCUU-CAGGCACAA .(((.(((((.((.......((-----(((.(((((...((--........)).))--)))))))).....-.....)).)))))((((((((.....))))))))..-..))).... ( -43.63, z-score = -2.74, R) >droSec1.super_10 2262894 109 + 3036183 AGCCAGGAGCUGAUAGAGCAUC-----CGGAGCACCACACACAUAUUGACCUGGGG--UGCCUGGACCAAC-CAACUUCCUUUCCGCUCGGCGGCAAUCGCUGAGCUU-CAGGCACAA .((((((((((.....))).((-----(((.(((((...((..........)).))--)))))))).....-.....))))....((((((((.....))))))))..-..))).... ( -41.60, z-score = -2.54, R) >droYak2.chrX 15913538 111 - 21770863 AGCCGGGAGCUGAUAGAGCGUCACCACCGGAGCAGCACGUAU-UGUGUGUUUGGGGG-UGCCUGGACCAAA----CUUCCGUUCCGCUCGGCACCAAUCGCCGAGCAA-CAGGCACAA .(((.(((((......((.(.((((.((.(((((.(((....-.)))))))).))))-)))))(((.....----..))))))))(((((((.......)))))))..-..))).... ( -47.50, z-score = -2.22, R) >droEre2.scaffold_4644 2442443 103 + 2521924 AGCCGGCAGCUGAUAGAGGAUA-----CGGAGCACCACUUA----UUGACUUGGGGGGUGCCUGGACCAAC-----UUCCGUUCCGCUCGGCACGAAUCGCUGAGCUU-CAGGCACAA .(((((.(((.((....((...-----(((.(((((.((((----......)))).))))))))..))...-----.)).)))))(((((((.......)))))))..-..))).... ( -39.50, z-score = -1.78, R) >consensus AGCCAGGAGCUGAUAGAGCAUC_____CGGAGCACCACAUA__UAUUGACCUGGGGG_UGCCUGGACCAAC_CAACUUCCGUUCCGCUCGGCGGCAAUCGCUGAGCUU_CAGGCACAA .(((.(((((.((...............((....))..............(..((.....))..)............)).)))))((((((((.....)))))))).....))).... (-27.98 = -28.78 + 0.80)



| Location | 2,489,907 – 2,490,011 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Shannon entropy | 0.30052 |
| G+C content | 0.54491 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2489907 104 + 22422827 -----CAUCCGGUGCACCACAU--AUAUUGACCUGGGGG-UGCCUGGACCAAAACAACUUCCGUUCCUCUCGGCGGCAAUCGCUGAGCUUUCAGGCACAAACAA-CUUACAAA -----..(((((.(((((.((.--.........))..))-))))))))..............((.((((((((((.....))))))).....))).))......-........ ( -28.90, z-score = -1.31, R) >droSim1.chrX 1766320 102 + 17042790 -----CAUCCGGAGCACCACAC--AUAUUGACCUGGGG--UGCCUGGACCAAC-CAACUUCCGUUCCGCUCGGCGGCAAUCGCUGAGCUU-CAGGCACAAACAAACUUACAAA -----..(((((..((......--....))..)))))(--((((((((.....-.(((....)))..((((((((.....))))))))))-)))))))............... ( -33.20, z-score = -2.69, R) >droSec1.super_10 2262912 104 + 3036183 -----CAUCCGGAGCACCACACACAUAUUGACCUGGGG--UGCCUGGACCAAC-CAACUUCCUUUCCGCUCGGCGGCAAUCGCUGAGCUU-CAGGCACAAACAAACUUACAAA -----..(((((..((............))..)))))(--((((((((.....-.............((((((((.....))))))))))-)))))))............... ( -31.16, z-score = -2.52, R) >droYak2.chrX 15913556 105 - 21770863 CGUCACCACCGGAGCAGCACGU--AUUGUGUGUUUGGGGGUGCCUGGACCAAA----CUUCCGUUCCGCUCGGCACCAAUCGCCGAGCAA-CAGGCACAAACAA-CUUACAAC ........((.(((((.(((..--...)))))))).)).(((((((.....((----(....)))..(((((((.......)))))))..-)))))))......-........ ( -38.30, z-score = -2.19, R) >droEre2.scaffold_4644 2442461 97 + 2521924 -----GAUACGGAGCACCACUU----AUUGACUUGGGGGGUGCCUGGACCAAC-----UUCCGUUCCGCUCGGCACGAAUCGCUGAGCUU-CAGGCACAAACAA-CUUACAAC -----......(((..((.(((----(......))))))(((((((((..(((-----....)))..(((((((.......)))))))))-)))))))......-)))..... ( -30.10, z-score = -2.20, R) >consensus _____CAUCCGGAGCACCACAU__AUAUUGACCUGGGGG_UGCCUGGACCAAC_CAACUUCCGUUCCGCUCGGCGGCAAUCGCUGAGCUU_CAGGCACAAACAA_CUUACAAA ........((((..((............))..))))....((((((((...........))).....((((((((.....))))))))....)))))................ (-20.84 = -21.44 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:35 2011