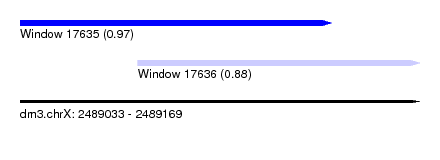
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,489,033 – 2,489,169 |
| Length | 136 |
| Max. P | 0.974795 |

| Location | 2,489,033 – 2,489,139 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.49850 |
| G+C content | 0.41682 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
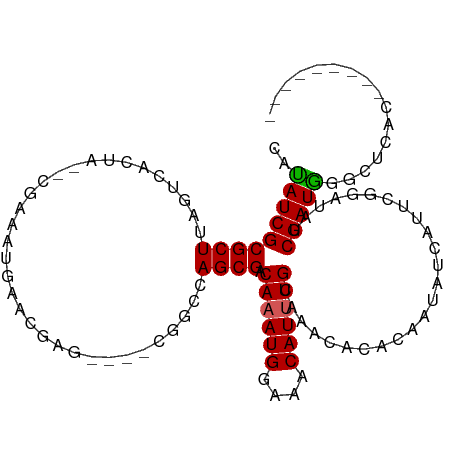
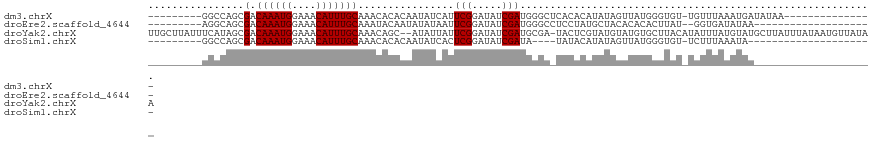
>dm3.chrX 2489033 106 + 22422827 CAUAUCGCGCUUAGUCACUAGUCACAAUGAACGAGCGAACGGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCAUUCGGAUAUCGAUGGGCUCACACAU----- .......(((((..(((..........)))..)))))...((((..(((((((((....))))))...........((((.....)))))))...)))).......----- ( -25.00, z-score = -2.16, R) >droEre2.scaffold_4644 2441649 101 + 2521924 CACAUCGCGCUUAGUCACUA--CGAUAUAAACAAG----CAGGCAGCGACAAAUGGAAACAUUUGCAAAUACAAUAUAUAAUUCGGAUAUCGAUGGGCCUCCUAUGC---- ........((.(((...(((--((((((...(..(----(.....))(.((((((....)))))))..................).)))))).))).....))).))---- ( -19.50, z-score = -0.73, R) >droYak2.chrX 15912738 106 - 21770863 CAUAUCGCGCUUAGUCACUG--UGAAAUGCUUGCUU-AUUUCAUAGCGACAAAUGGAAACAUUUGCAAA--CAGCAUAUUAUUCGGAUAUCGAUGCGAUACUCGUAUGUAU ..(((((((((..(((.(((--(((((((......)-))))))))).)))(((((....))))).....--.))).......(((.....))).))))))........... ( -31.40, z-score = -2.98, R) >droSec1.super_10 2262086 88 + 3036183 CAUAUCGCGCU-AGUCACGA-----AAUGAACGAG----CGGGAAGCGACAAAUGGAAACAUGUGCAAACACACAAUAUCAUUCGGAUAUCGAUGUAU------------- ....((.((((-..(((...-----..)))...))----)).)).(((....(((....))).)))....((((.(((((.....))))).).)))..------------- ( -21.10, z-score = -1.64, R) >droSim1.chrX 1765474 96 + 17042790 CAUAUCGCGCUUAGUCACUA--UGAAAUGAACGAG----CGGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCACUCGGAUAUCGAUAUAUACAU--------- .(((((((((((..(((...--.....)))..)))----))......(.((((((....))))))).........(((((.....)))))))))))......--------- ( -22.60, z-score = -2.88, R) >consensus CAUAUCGCGCUUAGUCACUA__CGAAAUGAACGAG____CGGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCAUUCGGAUAUCGAUGGGCUCAC_________ ..(((((((((.................................)))).((((((....)))))).........................)))))................ (-12.97 = -12.85 + -0.12)


| Location | 2,489,073 – 2,489,169 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 60.16 |
| Shannon entropy | 0.58731 |
| G+C content | 0.34956 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -9.18 |
| Energy contribution | -9.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2489073 96 + 22422827 ---------GGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCAUUCGGAUAUCGAUGGGCUCACACAUAUAGUUAUGGGUGU-UGUUUAAAUGAUAUAA--------------- ---------((((..(((((((((....))))))...........((((.....)))))))...))))(((.((((....)))).))).-................--------------- ( -22.20, z-score = -1.77, R) >droEre2.scaffold_4644 2441683 90 + 2521924 ---------AGGCAGCGACAAAUGGAAACAUUUGCAAAUACAAUAUAUAAUUCGGAUAUCGAUGGGCCUCCUAUGCUACACACACUUAU--GGUGAUAUAA-------------------- ---------.((((..(.((((((....)))))))..................(((..((....))..)))..)))).....(((....--.)))......-------------------- ( -17.20, z-score = -0.93, R) >droYak2.chrX 15912766 118 - 21770863 UUGCUUAUUUCAUAGCGACAAAUGGAAACAUUUGCAAACAGC--AUAUUAUUCGGAUAUCGAUGCGA-UACUCGUAUGUAUGUGCUUACAUAUUUAUGUAUGCUUAUUUAUAAUGUUAUAA (((((........)))))((((((....))))))..((((((--((((....((..(((((...)))-))..))...))))))((.(((((....))))).))..........)))).... ( -29.70, z-score = -2.10, R) >droSim1.chrX 1765508 86 + 17042790 ---------GGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCACUCGGAUAUCGAUA----UAUACAUAUAGUUAUGGGUGU-UCUUUAAAUA--------------------- ---------.......(.((((((....))))))).(((((.(.(((((.....))))).)...----....((((....)))).))))-).........--------------------- ( -16.90, z-score = -1.96, R) >consensus _________GGCCAGCGACAAAUGGAAACAUUUGCAAACACACAAUAUCAUUCGGAUAUCGAUGGGC_UACACAUAUAGAUAUGCGUAU_UGUUUAAAUAA____________________ ................(.((((((....)))))))................(((.....)))........................................................... ( -9.18 = -9.17 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:33 2011