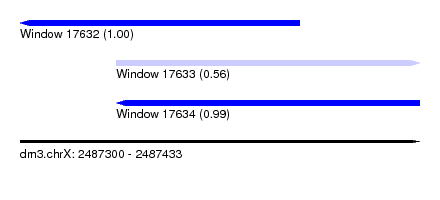
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,487,300 – 2,487,433 |
| Length | 133 |
| Max. P | 0.997559 |

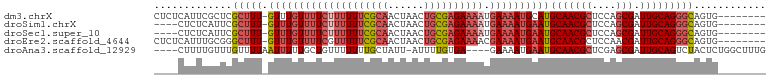
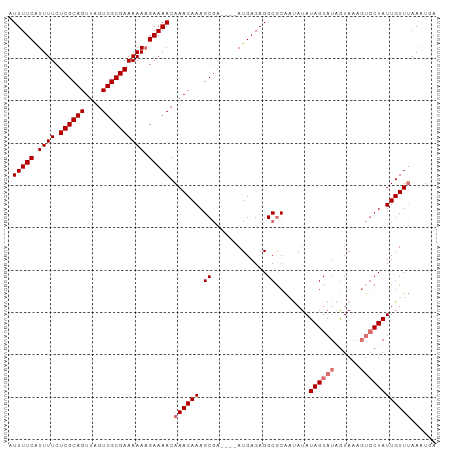
| Location | 2,487,300 – 2,487,393 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Shannon entropy | 0.36735 |
| G+C content | 0.42025 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -20.10 |
| Energy contribution | -22.46 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2487300 93 - 22422827 CUCUCAUUCGCUCGCUUU-GUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGCAUGCAACGCUCCAGCGAUUGCAGGGCAGUG-------- ....((((.((((....(-((..((((((((((((((((......)))))))))).)))))))))((((((((....))).)))))))))))))-------- ( -32.10, z-score = -3.32, R) >droSim1.chrX 1763759 89 - 17042790 ----CUCUCAUUCGCUUU-GUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUG-------- ----....((((.(((((-(((..(((((((((((((((......)))))))))).)))))..)))(((((((....))).)))))))))))))-------- ( -32.00, z-score = -4.08, R) >droSec1.super_10 2260436 89 - 3036183 ----CUCUCAUUCGCUUU-GUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUG-------- ----....((((.(((((-(((..(((((((((((((((......)))))))))).)))))..)))(((((((....))).)))))))))))))-------- ( -32.00, z-score = -4.08, R) >droEre2.scaffold_4644 2439846 93 - 2521924 CUCUCAUUUGCGGGCUUU-GUUUGUUUUCGUUUUUCGCAACUAACUGCGAGAAAACGAAAAUGAAUGCAACGCUCCAACGAUUGCAGGGCAGUG-------- ((((((.(((.((((.((-(((..(((((((((((((((......)))))).)))))))))..)..)))).))))...))).)).)))).....-------- ( -30.70, z-score = -2.81, R) >droAna3.scaffold_12929 2412637 93 - 3277472 ----CUUUUGUUUGUUUUAAUUUUUGCUGUUUUUUUGCUAUU-AUUUUGUGA----GAAAAUGAAUGCAACGCUCGAGCGAUUGCAGUCUACUCUGGCUUUG ----.....................((((((((((..(....-.....)..)----))))))((.((((((((....))).))))).))......))).... ( -19.10, z-score = -1.21, R) >consensus ____CUUUCGUUCGCUUU_GUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUG________ .............(((((.((((((((((((((((((((......)))))))))).))))))))))(((((((....))).)))))))))............ (-20.10 = -22.46 + 2.36)
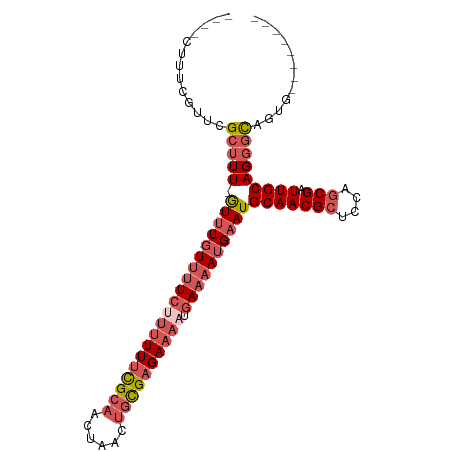

| Location | 2,487,332 – 2,487,433 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.20278 |
| G+C content | 0.32327 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -14.72 |
| Energy contribution | -15.72 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2487332 101 + 22422827 AUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGCGAGCGAAUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUUGUUUAAUUAA .(((((.((((.((((((....)))))).))))))))).((((((.(((..(......)...)))......((((((......))))))))))))...... ( -22.10, z-score = -1.62, R) >droSim1.chrX 1763791 97 + 17042790 AUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGCGA----AUGAGAGGCGCGAUAUAUAGUAUAGUAAAGUGCUAUUGUUUAAAUGA .(((((.((((.((((((....)))))).))))))))).((((((.(((.----.(....).)))......((((((......))))))))))))...... ( -20.60, z-score = -1.35, R) >droSec1.super_10 2260468 97 + 3036183 AUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGCGA----AUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUUGUUUAAAUGA .(((((.((((.((((((....)))))).))))))))).((((((.(((.----.(....).)))......((((((......))))))))))))...... ( -20.60, z-score = -1.66, R) >droEre2.scaffold_4644 2439878 95 + 2521924 AUUUUCGUUUUCUCGCAGUUAGUUGCGAAAAACGAAAACAAACAAAGCCCGCAAAUGAGAGGCACC-CAUAUAGUAUUAUA-----CUAUUGUUGAAAUUA .((((((((((.((((((....))))))))))))))))..(((((.((((........).)))...-....(((((...))-----))))))))....... ( -23.90, z-score = -3.68, R) >consensus AUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGCGA____AUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUUGUUUAAAUGA .(((((.((((.((((((....)))))))))).))))).((((((.((.............))........((((((......))))))))))))...... (-14.72 = -15.72 + 1.00)

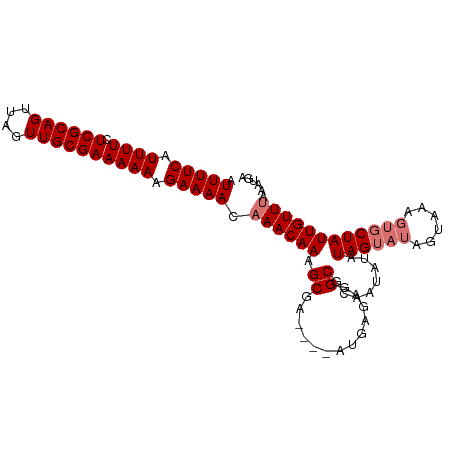
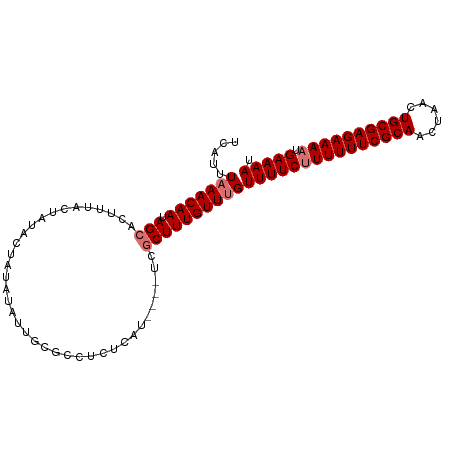
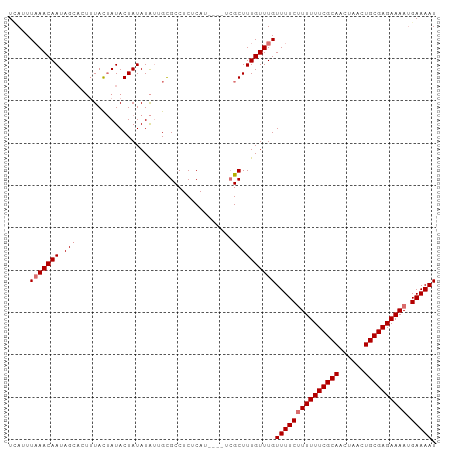
| Location | 2,487,332 – 2,487,433 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.20278 |
| G+C content | 0.32327 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2487332 101 - 22422827 UUAAUUAAACAAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAUUCGCUCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAU .....(((((((.(((.......((((...))))..(((.........)))..))))))))))(((((((((((((((......)))))))))).))))). ( -20.80, z-score = -3.43, R) >droSim1.chrX 1763791 97 - 17042790 UCAUUUAAACAAUAGCACUUUACUAUACUAUAUAUCGCGCCUCUCAU----UCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAU .....((((((((((.......))))..........(((........----.)))..))))))(((((((((((((((......)))))))))).))))). ( -18.60, z-score = -3.22, R) >droSec1.super_10 2260468 97 - 3036183 UCAUUUAAACAAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAU----UCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAU .....(((((((.(((...............................----..))))))))))(((((((((((((((......)))))))))).))))). ( -18.31, z-score = -3.06, R) >droEre2.scaffold_4644 2439878 95 - 2521924 UAAUUUCAACAAUAG-----UAUAAUACUAUAUG-GGUGCCUCUCAUUUGCGGGCUUUGUUUGUUUUCGUUUUUCGCAACUAACUGCGAGAAAACGAAAAU .......((((((((-----((...))))))...-...(((((......).))))..)))).((((((((((((((((......)))))).)))))))))) ( -23.70, z-score = -2.89, R) >consensus UCAUUUAAACAAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAU____UCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAU .....(((((((.(((.....................................))))))))))(((((((((((((((......)))))))))).))))). (-14.99 = -15.74 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:32 2011