
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,476,152 – 2,476,251 |
| Length | 99 |
| Max. P | 0.878135 |

| Location | 2,476,152 – 2,476,251 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.93 |
| Shannon entropy | 0.64275 |
| G+C content | 0.52469 |
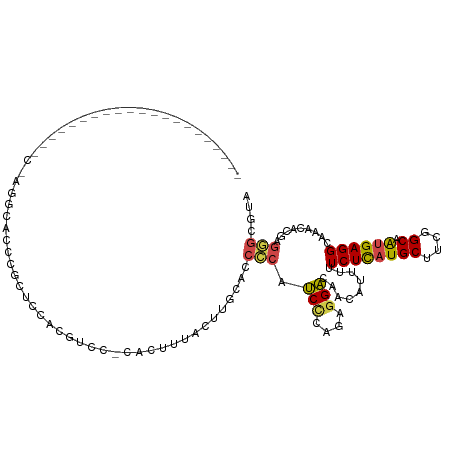
| Mean single sequence MFE | -27.01 |
| Consensus MFE | -9.67 |
| Energy contribution | -10.37 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
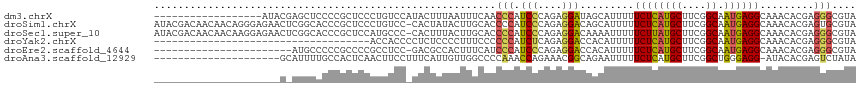
>dm3.chrX 2476152 99 + 22422827 ------------------AUACGAGCUCCCCGCUCCCUGUCCAUACUUUAAUUUCAACCCAUCCCAGAGGAUAGCAUUUUUCUCAUGCUUCGGCAAUGAGGCAAACACGAGGGCGUA ------------------.(((((((.....))))...((((..................((((....))))....(((.((((((((....)).)))))).))).....))))))) ( -25.00, z-score = -1.46, R) >droSim1.chrX 1752616 116 + 17042790 AUACGACAACAACAGGGAGAACUCGGCACCCGCUCCCUGUCC-CACUAUACUUGCACCCCAUCCCAGAGGACAGCAUUUUUCUCAUGCUUCGGCAAUGAGGCAAACACGAGUGCGUA .((((......((((((((....(((...)))))))))))..-((((....((((......(((....)))..........(((((((....)).))))))))).....)))))))) ( -35.00, z-score = -2.58, R) >droSec1.super_10 2249551 116 + 3036183 AUACGACAACAACAAGGAGAACUCGGCACCCGCUCCAUGCCC-CACUUUACUUGCACCCCAUCCCAGAGGACAAAAUUUUUCUUAUGCUUCGGCAAUGAGGCAAACACGAGGGCGUA ...............((((....(((...)))))))((((((-........((((......(((....)))..........(((((((....)).)))))))))......)))))). ( -27.54, z-score = -0.89, R) >droYak2.chrX 15899683 81 - 21770863 ------------------------------------ACCACCCCUCUCCCCUUUCCCCCCAUCUCAGAGGACCACAUUUUUCUCAUGCUUCGGCAAUGAGGCAAACACGAGGGCGUA ------------------------------------...(((((((...(((((...........)))))......(((.((((((((....)).)))))).)))...))))).)). ( -22.90, z-score = -2.59, R) >droEre2.scaffold_4644 2428621 93 + 2521924 -----------------------AUGCCCCCGCCCCGCCUCC-GACGCCACUUUCAUCCCAUCCCAGAGGACCACAUUUUUCUCAUGCUUCGGCAAUGAGGCAAACACGAGGGCGUA -----------------------((((((.((....(((((.-.......................(((((........))))).(((....)))..))))).....)).)))))). ( -28.70, z-score = -2.47, R) >droAna3.scaffold_12929 2402067 95 + 3277472 ---------------------GCAUUUUGCCACUCAACUUCCUUUCAUUGUUGGCCCCAAACCAGAAACGGCAGAAUUUUUCUCAUGCUUCGGCUGGGAGG-AUACACGAGUCUAUA ---------------------((.....)).((((..((.((((((....(((....)))....)))).)).))...((((((((.((....)))))))))-).....))))..... ( -22.90, z-score = -0.95, R) >consensus _____________________C_AGGCACCCGCUCCACGUCC_CACUUUACUUGCACCCCAUCCCAGAGGACAACAUUUUUCUCAUGCUUCGGCAAUGAGGCAAACACGAGGGCGUA .........................................................(((......(((((........))))).(((((((....))))))).......))).... ( -9.67 = -10.37 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:29 2011