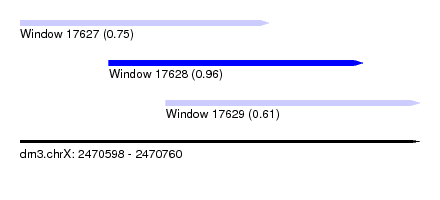
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,470,598 – 2,470,760 |
| Length | 162 |
| Max. P | 0.963196 |

| Location | 2,470,598 – 2,470,699 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Shannon entropy | 0.26718 |
| G+C content | 0.48457 |
| Mean single sequence MFE | -15.89 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.79 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
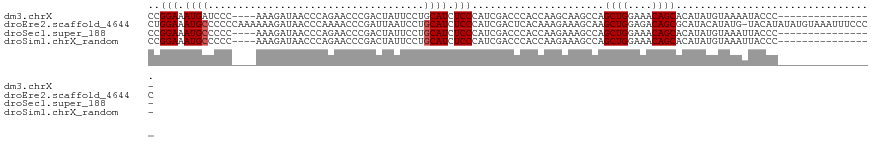
>dm3.chrX 2470598 101 + 22422827 CCGGAAAUGAUCCC----AAAGAUAACCCAGAACCCGACUAUUCCUGCAUCUCCCAUCGACCCACCAAGCAAGCCAGCUGGAAACAGCACAUAUGUAAAAUACCC---------------- ..(((.((((((..----...)))....(((.............)))))).)))......................((((....)))).................---------------- ( -13.42, z-score = -0.90, R) >droEre2.scaffold_4644 2422933 120 + 2521924 CUGGAAAUGCCCCCCAAAAAAGAUAACCCAAAACCCGAUUAAUCCUGCAUCUCCCAUCGACUCACAAAGAAAGCAAGCUGGAGACAGCGCAUACAUAUG-UACAUAUAUGUAAAUUUCCCC ..((((((((...................................(((.(((...............)))..))).((((....)))))).((((((((-....)))))))).)))))).. ( -22.06, z-score = -2.81, R) >droSec1.super_188 31365 101 + 32237 CCGGAAAUGCCCCC----AAAGAUAACCCAGAACCCGACUAUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUAUGUAAAUUACCC---------------- ..(((.((((....----............................)))).)))......................((((....)))).................---------------- ( -14.05, z-score = -1.33, R) >droSim1.chrX_random 1289510 101 + 5698898 CCGGAAAUGCCCCC----AAAGAUAACCCAGAACCCGACUAUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUAUGUAAAUUACCC---------------- ..(((.((((....----............................)))).)))......................((((....)))).................---------------- ( -14.05, z-score = -1.33, R) >consensus CCGGAAAUGCCCCC____AAAGAUAACCCAGAACCCGACUAUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUAUGUAAAUUACCC________________ ..(((.((((....................................)))).)))......................((((....))))................................. (-12.55 = -12.79 + 0.25)
| Location | 2,470,634 – 2,470,737 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Shannon entropy | 0.22208 |
| G+C content | 0.41899 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
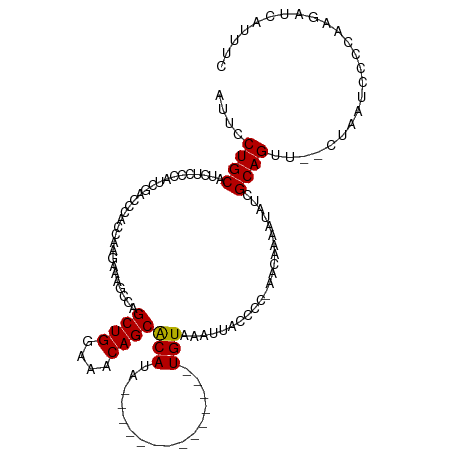
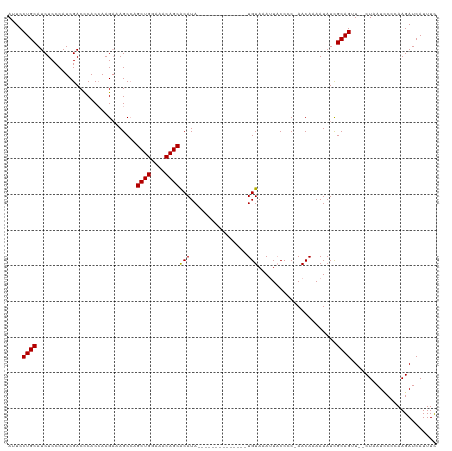
>dm3.chrX 2470634 103 + 22422827 AUUCCUGCAUCUCCCAUCGACCCACCAAGCAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAAUACCCC-AACAAAAUAUCGCAGUU--CUUAUACCCAAGAUCAUUUU ....((((............................((((....))))((...--------------.))..........-...........)))).(--(((......))))....... ( -14.70, z-score = -2.23, R) >droEre2.scaffold_4644 2422973 120 + 2521924 AAUCCUGCAUCUCCCAUCGACUCACAAAGAAAGCAAGCUGGAGACAGCGCAUACAUAUGUACAUAUAUGUAAAUUUCCCCCAACAAAAUAUUGCAGAUAUCUAAUUCCUAAGAUCAUUUC ....(((((.......................((..((((....)))))).((((((((....))))))))....................)))))..((((........))))...... ( -22.40, z-score = -2.71, R) >droSec1.super_188 31401 103 + 32237 AUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAUUACCCC-AACAAAAUAUCGCAGUU--CUAAUCCCCAAGAUCAUUUC ....((((............................((((....))))((...--------------.))..........-...........))))..--.................... ( -14.10, z-score = -2.01, R) >droSim1.chrX_random 1289546 103 + 5698898 AUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAUUACCCC-AACAAAAUAUCGCAGUU--CUAAUCCCCAAGAUCAUUUC ....((((............................((((....))))((...--------------.))..........-...........))))..--.................... ( -14.10, z-score = -2.01, R) >consensus AUUCCUGCAUCUCCCAUCGACCCACCAAGAAAGCCAGCUGGAAACAGCACAUA______________UGUAAAUUACCCC_AACAAAAUAUCGCAGUU__CUAAUCCCCAAGAUCAUUUC ....((((............................((((....))))...................(((............))).......))))........................ (-13.73 = -13.72 + -0.00)

| Location | 2,470,657 – 2,470,760 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Shannon entropy | 0.22208 |
| G+C content | 0.37842 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.33 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
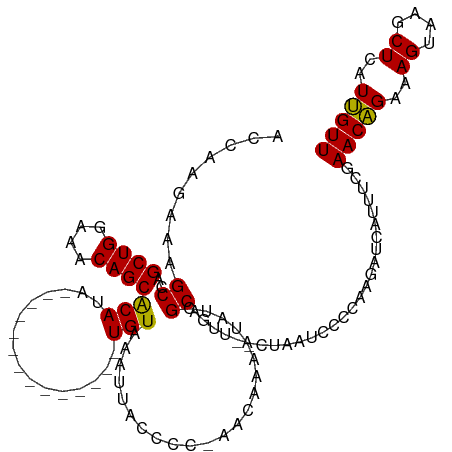
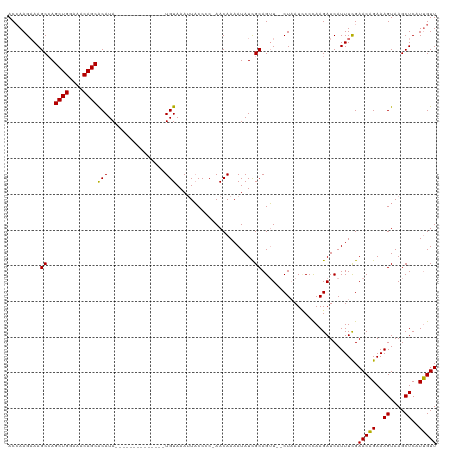
>dm3.chrX 2470657 103 + 22422827 ACCAAGCAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAAUACCCC-AACAAAAUAUCGCAGUU--CUUAUACCCAAGAUCAUUUUGAACAGAAAGUAAGCUCAUUGUU ..(((...(((..((((....)))).....--------------.............-..(((((((((...((.--.....))....))).))))))............)))..))).. ( -17.00, z-score = -1.73, R) >droEre2.scaffold_4644 2422996 120 + 2521924 ACAAAGAAAGCAAGCUGGAGACAGCGCAUACAUAUGUACAUAUAUGUAAAUUUCCCCCAACAAAAUAUUGCAGAUAUCUAAUUCCUAAGAUCAUUUCGAACCGAAAGUAAGCUCAUGGUU .....((((((((((((....))))...((((((((....))))))))...................))))....((((........))))..)))).(((((..((....))..))))) ( -25.50, z-score = -2.72, R) >droSec1.super_188 31424 103 + 32237 ACCAAGAAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAUUACCCC-AACAAAAUAUCGCAGUU--CUAAUCCCCAAGAUCAUUUCGAACAGAAAGUAAGCUCAUUGUU ..(((...(((..((((....)))).....--------------......((((...-..............(.(--((........))).).((((.....)))))))))))..))).. ( -15.60, z-score = -1.77, R) >droSim1.chrX_random 1289569 103 + 5698898 ACCAAGAAAGCCAGCUGGAAACAGCACAUA--------------UGUAAAUUACCCC-AACAAAAUAUCGCAGUU--CUAAUCCCCAAGAUCAUUUCGAACAGAAAGUAAGCUCAUUGUU ..(((...(((..((((....)))).....--------------......((((...-..............(.(--((........))).).((((.....)))))))))))..))).. ( -15.60, z-score = -1.77, R) >consensus ACCAAGAAAGCCAGCUGGAAACAGCACAUA______________UGUAAAUUACCCC_AACAAAAUAUCGCAGUU__CUAAUCCCCAAGAUCAUUUCGAACAGAAAGUAAGCUCAUUGUU .........((..((((....))))...................(((............))).......))...........................(((((..((....))..))))) (-12.70 = -12.33 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:28 2011