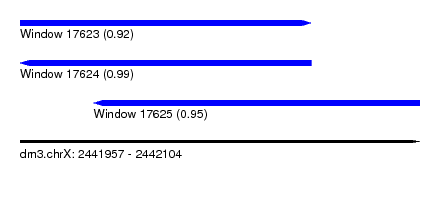
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,441,957 – 2,442,104 |
| Length | 147 |
| Max. P | 0.992765 |

| Location | 2,441,957 – 2,442,064 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.61 |
| Shannon entropy | 0.56477 |
| G+C content | 0.41270 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -12.30 |
| Energy contribution | -16.43 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2441957 107 + 22422827 ------------AAUACACAGCGAAACAGCUUAUU-GGGGUAAAGUUUGCAAAAAAUAUAACUCAUACGCCCAGUGUAACAGGUCAAGUUGUGUUCAACAUGGCGUAUGAGCGAUAUUCC ------------........(((((...((((...-.))))....)))))....(((((..((((((((((..((((((((.(......).))))..))))))))))))))..))))).. ( -29.70, z-score = -2.59, R) >droSec1.super_10 2215583 93 + 3036183 -----------------------GAACAGCUUAUU-GUAGUAAAGUUGGCAG---AUAUUACCCAUACGCCCAGUGUUACAGGUCAAGUUGUGUUCAACAUGGCGUAUGAGCGAUAUUCC -----------------------...((((((...-......))))))...(---((((..(.((((((((..((((((((.(......).)))..))))))))))))).)..))))).. ( -25.10, z-score = -1.97, R) >droYak2.chrX 15868011 117 - 21770863 AAUAUCUGGGAAAACAGCUUAUUGUGCAGCUAUCUUGGAGUAAAGCUUGCAG---AUAUAACUCAUCCGCCCCGUAUUGCUGAUGAAGUUUUGUUCAACGUGGCGUAUAAGCGAUAUUCC ((((((((......))(((...(((((.(((((.(((((.(((((((.....---.......(((((.((........)).))))))))))))))))).))))))))))))))))))).. ( -28.20, z-score = -0.48, R) >droEre2.scaffold_4644 2392346 110 + 2521924 AAUAACUAUGGAAAUAGCCUAUUGGGCAGCAAACUAAGAGUAAUGUUUGCAA---AUAUAACUCAUACGCCCAGUGUGGCUGGUGAUGUAUCAUC-------GCGUAUAUGCGAUAUUCC .........((((.(((((((((((((.((((((..........))))))..---.(((.....))).)))))))).)))))..........(((-------(((....)))))).)))) ( -34.00, z-score = -2.86, R) >consensus ____________AAUAGCCUAUUGAACAGCUAACU_GGAGUAAAGUUUGCAA___AUAUAACUCAUACGCCCAGUGUUACAGGUCAAGUUGUGUUCAACAUGGCGUAUAAGCGAUAUUCC ............................(((((((........))))))).....((((..((((((((((......((((((......))))))......))))))))))..))))... (-12.30 = -16.43 + 4.12)

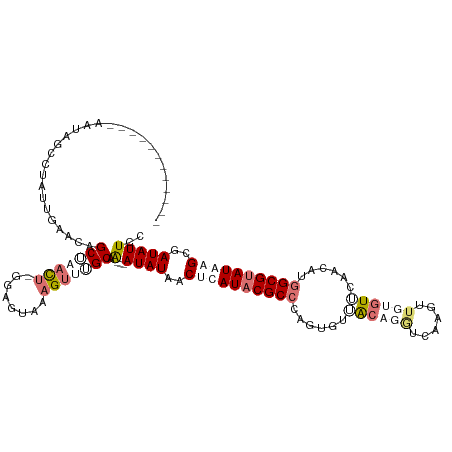
| Location | 2,441,957 – 2,442,064 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.61 |
| Shannon entropy | 0.56477 |
| G+C content | 0.41270 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.91 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2441957 107 - 22422827 GGAAUAUCGCUCAUACGCCAUGUUGAACACAACUUGACCUGUUACACUGGGCGUAUGAGUUAUAUUUUUUGCAAACUUUACCCC-AAUAAGCUGUUUCGCUGUGUAUU------------ (((((((.(((((((((((......((((..........))))......))))))))))).)))))))...............(-(...(((......))).))....------------ ( -27.20, z-score = -2.28, R) >droSec1.super_10 2215583 93 - 3036183 GGAAUAUCGCUCAUACGCCAUGUUGAACACAACUUGACCUGUAACACUGGGCGUAUGGGUAAUAU---CUGCCAACUUUACUAC-AAUAAGCUGUUC----------------------- ((.((((..((((((((((.(((((....(.....).....)))))...))))))))))..))))---...))...........-............----------------------- ( -24.70, z-score = -2.59, R) >droYak2.chrX 15868011 117 + 21770863 GGAAUAUCGCUUAUACGCCACGUUGAACAAAACUUCAUCAGCAAUACGGGGCGGAUGAGUUAUAU---CUGCAAGCUUUACUCCAAGAUAGCUGCACAAUAAGCUGUUUUCCCAGAUAUU ..((((((((......))...(((((...........))))).....((((((((((.....)))---))))............(((((((((........)))))))))))).)))))) ( -28.60, z-score = -1.55, R) >droEre2.scaffold_4644 2392346 110 - 2521924 GGAAUAUCGCAUAUACGC-------GAUGAUACAUCACCAGCCACACUGGGCGUAUGAGUUAUAU---UUGCAAACAUUACUCUUAGUUUGCUGCCCAAUAGGCUAUUUCCAUAGUUAUU ((((((((((......))-------))))..........((((....((((((((.....)))..---..((((((..........)))))).)))))...))))..))))......... ( -30.50, z-score = -2.89, R) >consensus GGAAUAUCGCUCAUACGCCAUGUUGAACAAAACUUCACCAGCAACACUGGGCGUAUGAGUUAUAU___CUGCAAACUUUACUCC_AAUAAGCUGCUCAAUAGGCUAUU____________ ...((((.(((((((((((..............................))))))))))).))))....................................................... (-13.53 = -13.91 + 0.38)

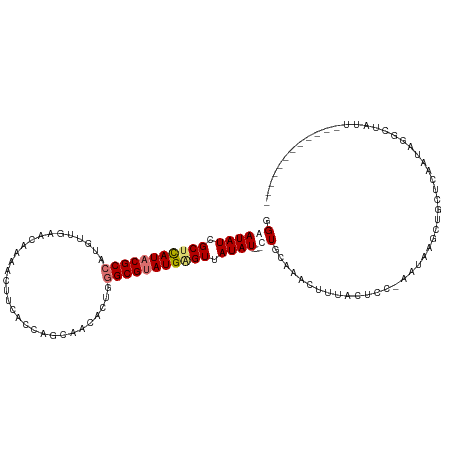
| Location | 2,441,984 – 2,442,104 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Shannon entropy | 0.35690 |
| G+C content | 0.41693 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -20.13 |
| Energy contribution | -21.44 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
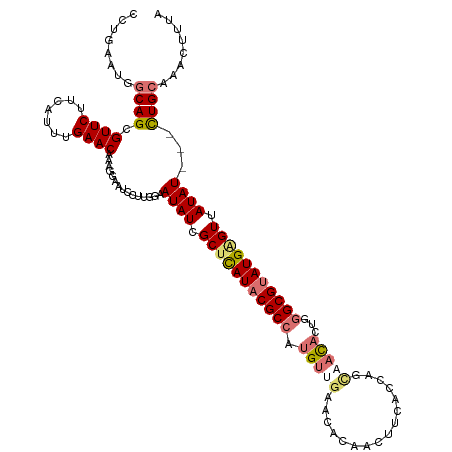
>dm3.chrX 2441984 120 - 22422827 CCUGAAUGGCAGCGUUCUUCUUUUGAACAAUGGAAACCUUGGAAUAUCGCUCAUACGCCAUGUUGAACACAACUUGACCUGUUACACUGGGCGUAUGAGUUAUAUUUUUUGCAAACUUUA ........((((.((((.......))))...(....)...(((((((.(((((((((((......((((..........))))......))))))))))).)))))))))))........ ( -32.00, z-score = -2.94, R) >droSec1.super_10 2215599 109 - 3036183 --------GCAGCGUUCUUCUUUCGAACAAAGGAAUCGUUGGAAUAUCGCUCAUACGCCAUGUUGAACACAACUUGACCUGUAACACUGGGCGUAUGGGUAAUAU---CUGCCAACUUUA --------.....((((.......)))).........(((((.((((..((((((((((.(((((....(.....).....)))))...))))))))))..))))---...))))).... ( -32.70, z-score = -3.02, R) >droYak2.chrX 15868051 117 + 21770863 CCUGAAUGGCAGCGUUCUUGAUUUGAACAAAGGAAUCCUUGGAAUAUCGCUUAUACGCCACGUUGAACAAAACUUCAUCAGCAAUACGGGGCGGAUGAGUUAUAU---CUGCAAGCUUUA ((((..(((((((((((.......)))).((((...))))........))).....)))).(((((...........)))))....))))(((((((.....)))---))))........ ( -27.90, z-score = -0.56, R) >droEre2.scaffold_4644 2392386 110 - 2521924 ACUGAAUGACAGCGUUCUCUAUGUGAACAAAGAAAUCCUUGGAAUAUCGCAUAUACGCGAUGAU-------ACAUCACCAGCCACACUGGGCGUAUGAGUUAUAU---UUGCAAACAUUA ....((((...((((((.......)))).............((((((.((.(((((((((((..-------.)))).((((.....))))))))))).)).))))---))))...)))). ( -28.00, z-score = -2.10, R) >consensus CCUGAAUGGCAGCGUUCUUCAUUUGAACAAAGGAAUCCUUGGAAUAUCGCUCAUACGCCAUGUUGAACACAACUUCACCAGCAACACUGGGCGUAUGAGUUAUAU___CUGCAAACUUUA ........((((.((((.......))))...............((((.(((((((((((.(((((................)))))...))))))))))).))))...))))........ (-20.13 = -21.44 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:25 2011