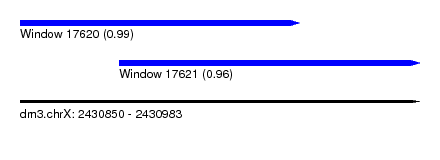
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,430,850 – 2,430,983 |
| Length | 133 |
| Max. P | 0.994105 |

| Location | 2,430,850 – 2,430,943 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.22232 |
| G+C content | 0.56810 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -30.22 |
| Energy contribution | -31.01 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
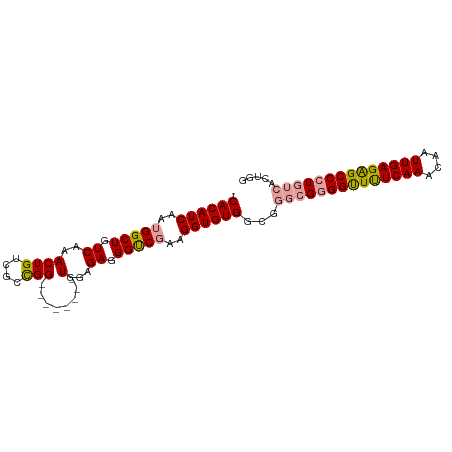
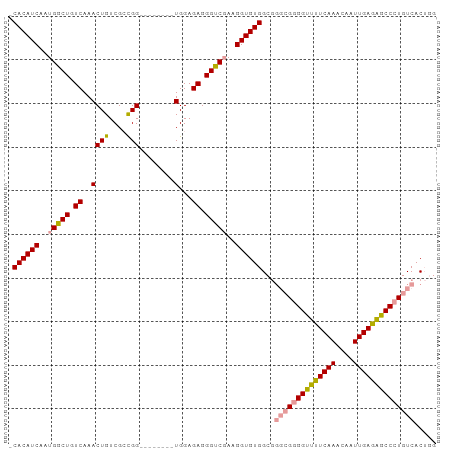
>dm3.chrX 2430850 93 + 22422827 -CACAUCAAUGGCUGUCAAACUGUCGAUGGAUGGAUGGUGGAGAGGGCCGAAGGUGUGGC----GCGGCUUUCAAACAAUUGAGAGCCCUGUCACAUG -........(((((.((..((((((........))))))...)).)))))...(((((((----(.(((((((((....))))))))).)))))))). ( -39.00, z-score = -3.54, R) >droSim1.chrX 1735601 90 + 17042790 GCACAUCAAUGGCUGUCAAACUGUCGCCGG--------UGGAGAGGGUCGAAGGUGUGGCGGGCGGGGUUCUCAAACAAUUGAGGGCCCUGUCACUGG .((((((..(((((.((..((((....)))--------)...)).)))))..))))))...((((((((((((((....))))))))))))))..... ( -40.80, z-score = -3.34, R) >droSec1.super_10 2205009 90 + 3036183 ACACAUCAAUGGCUGUCAAACUGUCGCCGG--------UGGAGAGGGUCCAAGGUGUGGCGGGCGGGGUUUUCAAACAAUUGAGAGCCCUGUCACUGG .((((((...((((.((..((((....)))--------)...)).))))...))))))...((((((((((((((....))))))))))))))..... ( -38.10, z-score = -2.74, R) >consensus _CACAUCAAUGGCUGUCAAACUGUCGCCGG________UGGAGAGGGUCGAAGGUGUGGCGGGCGGGGUUUUCAAACAAUUGAGAGCCCUGUCACUGG .((((((..(((((.((.........................)).)))))..))))))...((((((((((((((....))))))))))))))..... (-30.22 = -31.01 + 0.79)

| Location | 2,430,883 – 2,430,983 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Shannon entropy | 0.35458 |
| G+C content | 0.56872 |
| Mean single sequence MFE | -42.75 |
| Consensus MFE | -25.39 |
| Energy contribution | -27.70 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
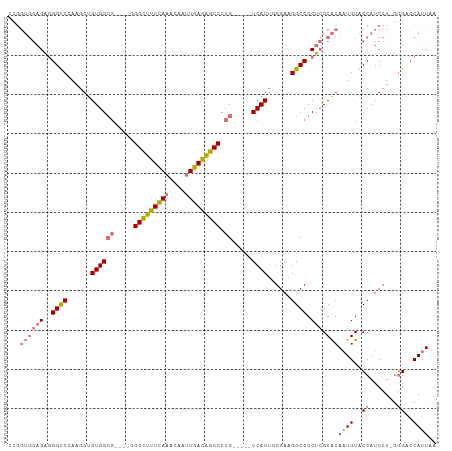
>dm3.chrX 2430883 100 + 22422827 AUGGUGGAGAGGGCCGAAGGUGUGGCG----CGGCUUUCAAACAAUUGAGAGCCCUG-----UCACAUGGAAGGCCGCUCGCACAUUGUACCAUCCUAGGGAACAUUAA ...((((((..((((....((((((((----.(((((((((....))))))))).))-----))))))....)))).))).)))..(((.((.......)).))).... ( -42.50, z-score = -3.38, R) >droSim1.chrX 1735627 103 + 17042790 CCGGUGGAGAGGGUCGAAGGUGUGGCGGGCGGGGUUCUCAAACAAUUGAGGGCCCUG-----UCACUGGGAAGGCCGCUCGCACAAUGUGCCAUCCU-GGGCCCAUUAA ..........(((((..(((.(((((.((((((((((((((....))))))))))))-----)).((....)))))))..((((...))))...)))-.)))))..... ( -46.60, z-score = -1.77, R) >droSec1.super_10 2205035 103 + 3036183 CCGGUGGAGAGGGUCCAAGGUGUGGCGGGCGGGGUUUUCAAACAAUUGAGAGCCCUG-----UCACUGGGAAGGCCGCUCGCACAAUGUGCCAUCCU-GGGACCAUUAA ...........(((((.(((.(((((.((((((((((((((....))))))))))))-----)).((....)))))))..((((...))))...)))-.)))))..... ( -45.90, z-score = -2.33, R) >droYak2.chrX 15856828 94 - 21770863 ------AUGGAGGUCCAAGGUGUGG-------GGCUUUUACACAACUAAGAGCCCCGCCUUUUCACUUGGAAGGCCGCUAGCGCAAUGUACCAUCCU--AGCCCAUUAA ------..((((((.((..((((((-------((((((((......))))))))))(((((((.....))))))).....))))..)).))).))).--.......... ( -36.00, z-score = -2.65, R) >consensus CCGGUGGAGAGGGUCCAAGGUGUGGCG____GGGCUUUCAAACAAUUGAGAGCCCUG_____UCACUGGGAAGGCCGCUCGCACAAUGUACCAUCCU_GGGACCAUUAA ...((((((..((((......((((....((((((((((((....)))))))))))).....))))......)))).))).)))((((..((.......))..)))).. (-25.39 = -27.70 + 2.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:22 2011