
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,782,338 – 9,782,423 |
| Length | 85 |
| Max. P | 0.692705 |

| Location | 9,782,338 – 9,782,423 |
|---|---|
| Length | 85 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Shannon entropy | 0.63239 |
| G+C content | 0.54031 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -9.50 |
| Energy contribution | -9.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9782338 85 + 23011544 CGAUUGCCGAGCUGU---UUGAUCACAAAACUGGCUGCCAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC---CGCUAGACCACGUAAC----- .....((.(..(((.---.((...((..((((((.((((....))))...)))))).))...))..)))..)---.)).............----- ( -19.80, z-score = -0.25, R) >droSim1.chr2L 9561250 84 + 22036055 CGAUUGCCGAGC----CGUUGAUCACAAAACUGGCUGCCAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC---CGCCAGACCACGUAAC----- .((((..((...----))..))))......(((((.(((..(((((((..........)))))))...).))---.)))))..........----- ( -23.60, z-score = -1.85, R) >droSec1.super_3 5229759 84 + 7220098 CGAUUGCCGAGC----CGUUGAUCACAAAACUGGCUGCUAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC---CGCCAGACCACGUAAC----- .((((..((...----))..))))......(((((.(((..(((((((..........)))))))....)))---.)))))..........----- ( -25.90, z-score = -2.59, R) >droYak2.chr2L 12470657 88 + 22324452 CGAUUGCCGAUCCAUCCGAUGAUCACAAAACUGGCCGCCAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC---AGCCAGACCACGUAAC----- .((((..((.......))..))))......(((((.(((..(((((((..........)))))))...).))---.)))))..........----- ( -25.00, z-score = -3.01, R) >droEre2.scaffold_4929 10394359 84 + 26641161 CAAUUGCCGAUG----CGAUGAUCACAAAACUGGCUGCCAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC---AGCCAGACCACGUAAC----- ..(((((....)----))))..........(((((((((..(((((((..........)))))))...).))---))))))..........----- ( -28.00, z-score = -3.72, R) >droAna3.scaffold_12943 1222050 83 + 5039921 CCAUCGGCGAUC-----GGCGAUCACAAAACCUGGUGCCAUAUGGCGAAACCAGUUAGUCGCCUCCCAGAGC---AGCCAGCGAACGCAAC----- ......(((.((-----((((((........((((((((....)))...)))))...)))))).......((---.....)))).)))...----- ( -24.90, z-score = -1.23, R) >droPer1.super_1 8417822 85 - 10282868 CGAGUGCGACUC----GUCAGAGCACAAAACUGGAUGCCAGAUGGCGAAACCAGUUAGUCGCCACCCAGAGCU-CCACCAGCGAACGCAA------ ....((((..((----((..((((....((((((.((((....))))...))))))..((........)))))-).....)))).)))).------ ( -25.00, z-score = -1.51, R) >dp4.chr4_group3 11302795 85 - 11692001 CGAGUGCGACUC----GUCAGAGUACAAAACUGGAUGCCAGAUGGCGAAACCAGUUAGUCGCCACCCAGAGCU-CCACCAGCGAACGCCA------ .(.(((((((((----....))......((((((.((((....))))...)))))).)))).))).)...(((-.....)))........------ ( -22.70, z-score = -0.88, R) >droWil1.scaffold_180772 8423005 87 + 8906247 ----CGAUCAAC-----AUUCAGCACAAAACUGGCUGCUAUAUGAGCAAACCAGUUAGUCGUCGUCGUCAGCUCAAAGCCGUAAGCAACACUUAAU ----........-----....(((....((((((.((((.....))))..)))))).(.((....)).).)))....((.....)).......... ( -14.90, z-score = -0.04, R) >droMoj3.scaffold_6500 3573686 87 - 32352404 CGAGCGCUGAGC------UUUUGUACAAAACUGGCCGUCGUUUGGAUAAACCAGUUAGUCGGCCGACAGAGUA-GCACGCGUGAACAGCUGGAC-- (.(((((.(.((------((((((.......((((((.(..((((.....))))...).)))))))))))..)-)).)))(....).))).)..-- ( -25.41, z-score = -0.11, R) >droVir3.scaffold_12963 18373766 87 - 20206255 UGAGUGCUUGGC------UUUAGUACAAAACUGGCCGUCGUAUGGCGAAACCAGUUAGUCGCCGCACAGAGUA-GCACGCGUAAACAGCUGAAU-- ...(((((.(((------(..(((.....)))))))...((.((((((..........)))))).)).....)-))))((.......)).....-- ( -23.20, z-score = 0.41, R) >consensus CGAUUGCCGAGC_____GUUGAUCACAAAACUGGCUGCCAUAUGGCGAAACCAGUUAGUCGCCAUCCAGAGC___CGCCAGACAACGUAAC_____ ............................((((((.((((....))))...))))))........................................ ( -9.50 = -9.74 + 0.24)

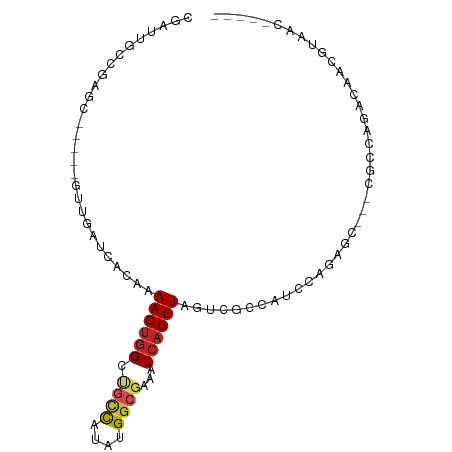
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:56 2011