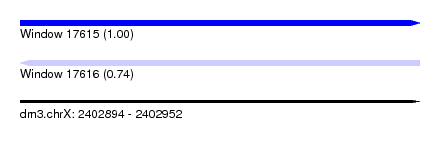
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,402,894 – 2,402,952 |
| Length | 58 |
| Max. P | 0.997652 |

| Location | 2,402,894 – 2,402,952 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.57516 |
| G+C content | 0.49978 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -10.03 |
| Energy contribution | -9.76 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
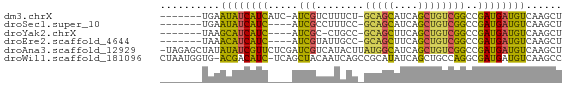
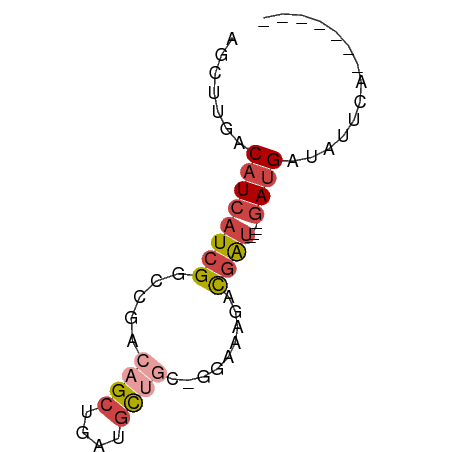
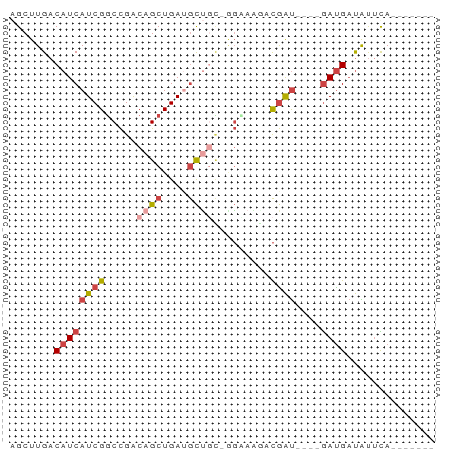
>dm3.chrX 2402894 58 + 22422827 -------UGAAUAUCAUCAUC-AUCGUCUUUCU-GCAGCAUCAGCUGUCGGCCGAUGAUGUCAAGCU -------..........((((-((((.((....-(((((....))))).)).))))))))....... ( -17.30, z-score = -1.90, R) >droSec1.super_10 2178072 55 + 3036183 -------UGAAUAUCAUC----AUCGCCUUUCC-GCAGCAUCAGCUGUCGGCCGAUGAUGUCAAGCU -------.......((((----((((.....((-(((((....)))).))).))))))))....... ( -19.10, z-score = -2.71, R) >droYak2.chrX 15828006 54 - 21770863 -------UAAGCAUCAUC----AUCGC-CUGCC-GCAGCUUCAGCUGUCGGCCGAUGAUGUCAAGCU -------..(((..((((----((((.-..(((-(((((....)))).))))))))))))....))) ( -25.80, z-score = -4.68, R) >droEre2.scaffold_4644 2352696 55 + 2521924 -------UAAACAUCAUC----AUCGUAUUGCC-GCAGCUUCAGCUGUCGGCCGAUGAUGUCAAGCU -------.......((((----((((....(((-(((((....)))).))))))))))))....... ( -23.10, z-score = -4.80, R) >droAna3.scaffold_12929 2313921 66 + 3277472 -UAGAGCUAUAUAUCGUUCUCGAUCGUCAUACUUAUGGCAUCAGCUGUCGGCCGAUGAUGUCAAGCU -...((((..((((((((...(((.(((((....)))))))).((.....)).))))))))..)))) ( -16.20, z-score = -0.32, R) >droWil1.scaffold_181096 12112089 65 + 12416693 CUAAUGGUG-ACGACAUC-UCAGCUACAAUCAGCCGCAUAUCAGCUGCCAGGCGAUGAUGUCAAGCC .....(((.-..((((((-...(((.....((((.........))))...)))...))))))..))) ( -17.60, z-score = -0.83, R) >consensus _______UAAACAUCAUC____AUCGCCUUGCC_GCAGCAUCAGCUGUCGGCCGAUGAUGUCAAGCU ..........((((((((.............((.(((((....))))).))..))))))))...... (-10.03 = -9.76 + -0.27)
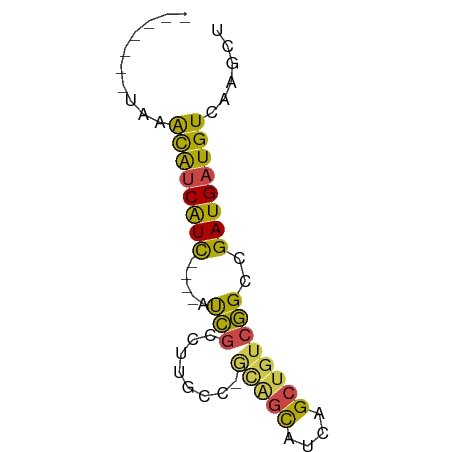

| Location | 2,402,894 – 2,402,952 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.57516 |
| G+C content | 0.49978 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -5.35 |
| Energy contribution | -6.02 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
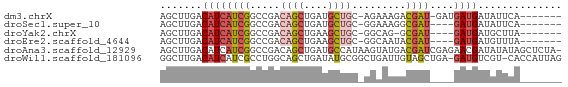
>dm3.chrX 2402894 58 - 22422827 AGCUUGACAUCAUCGGCCGACAGCUGAUGCUGC-AGAAAGACGAU-GAUGAUGAUAUUCA------- .......((((((((..(..((((....)))).-.).....))))-))))..........------- ( -17.00, z-score = -1.85, R) >droSec1.super_10 2178072 55 - 3036183 AGCUUGACAUCAUCGGCCGACAGCUGAUGCUGC-GGAAAGGCGAU----GAUGAUAUUCA------- .......((((((((.(((.((((....)))))-)).....))))----)))).......------- ( -21.60, z-score = -3.07, R) >droYak2.chrX 15828006 54 + 21770863 AGCUUGACAUCAUCGGCCGACAGCUGAAGCUGC-GGCAG-GCGAU----GAUGAUGCUUA------- (((....((((((((((((.((((....)))))-)))..-.))))----))))..)))..------- ( -25.60, z-score = -3.88, R) >droEre2.scaffold_4644 2352696 55 - 2521924 AGCUUGACAUCAUCGGCCGACAGCUGAAGCUGC-GGCAAUACGAU----GAUGAUGUUUA------- .......((((((((((((.((((....)))))-)))....))))----)))).......------- ( -24.00, z-score = -4.52, R) >droAna3.scaffold_12929 2313921 66 - 3277472 AGCUUGACAUCAUCGGCCGACAGCUGAUGCCAUAAGUAUGACGAUCGAGAACGAUAUAUAGCUCUA- .(((((....(((((((.....)))))))...)))))......((((....))))...........- ( -13.50, z-score = -0.14, R) >droWil1.scaffold_181096 12112089 65 - 12416693 GGCUUGACAUCAUCGCCUGGCAGCUGAUAUGCGGCUGAUUGUAGCUGA-GAUGUCGU-CACCAUUAG ((..(((((((.(((.(((.((((((.....))))))....))).)))-))))))).-..))..... ( -22.30, z-score = -1.40, R) >consensus AGCUUGACAUCAUCGGCCGACAGCUGAUGCUGC_GGAAAGACGAU____GAUGAUAUUCA_______ ..........(((((((.....)))))))...................................... ( -5.35 = -6.02 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:18 2011