| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,769,030 – 9,769,125 |
| Length | 95 |
| Max. P | 0.748383 |

| Location | 9,769,030 – 9,769,120 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.72 |
| Shannon entropy | 0.70744 |
| G+C content | 0.49504 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -8.51 |
| Energy contribution | -8.84 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
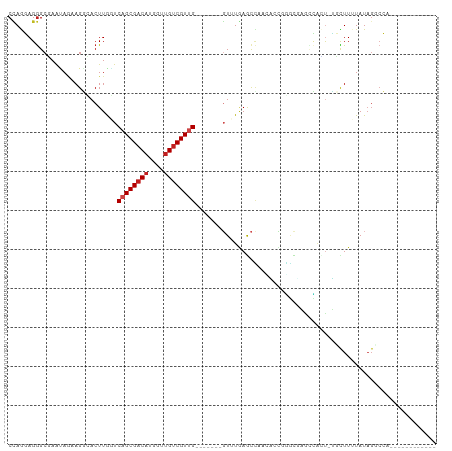
>dm3.chr2L 9769030 90 - 23011544 CCAGGAGGGCGAAUAGAAGGCACUUGUUCAGCGACAUCGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCCUUUUAUAGGCCU------------ ......((((..((((((((.........(((((((....)))))))(-------((...((((.......)))).)))...-.))))))))..))))------------ ( -30.40, z-score = -1.85, R) >droVir3.scaffold_12963 18358104 101 + 20206255 CCAACAGGGCAAAGAGAAGCAACUUAUUCAGCGACAUGGCGGUCGUUG-------GUAUAAACAAUACGAUUUCGCGUGGUU-GGUUUAUAUAGUCGAUGG-GGUUUUGG ((((....((........))..(((((((((((((......)))))))-------((((((((..((((......))))...-.))))))))....)))))-)...)))) ( -25.30, z-score = -1.36, R) >droGri2.scaffold_14978 474892 102 - 1124632 CCAACAGGGCUAACAGUAGCGACUUGUUCAGCGACAUGGUUGUCGUUG-------UUCUAAAUUAGACAAUCGUGUUAGAUU-GGCUUAUAUAGUCGAUGGCUGUUGAUA .((((((.(((((((((....)).)))).)))((((((((((((....-------..........))))))))))))..(((-((((.....)))))))..))))))... ( -33.14, z-score = -2.90, R) >droMoj3.scaffold_6500 3543996 101 + 32352404 CAAACAGAGCGAAAAGAAGCGACUUGUUCAGCGACAUGGUUGUCGUGG-------UUAUUAGCCAUUUAAGGUCGCGUGGUU-GGUUUUUAUUGUCGCUAG-AGCUUAUA .......(((((.(((((.(((((......((((((((((((......-------....))))))).....)))))..))))-).)))))....)))))..-........ ( -26.90, z-score = -0.86, R) >droWil1.scaffold_180772 1141548 108 + 8906247 CCAAAAGGGCCAAAACUAGAGACUUAUUCAGCGACAUGACUGCCGUUGGUAGUACGUAUGAGCUCUUAACGAACUUGUCAACAUGCUUUUAUACCAAUUUACUGCUCU-- .....(((((.......((((.(((((...(((.....((((((...)))))).))))))))))))....((.....))........................)))))-- ( -18.20, z-score = 0.81, R) >droPer1.super_1 8405487 101 + 10282868 CCAGGAGGGCCAACAAAAGGGACUUGGUCAGCGACAUAGUUGUCGUUG----UUUGUAUGAGCUCCAAACGGGCGAGAUGGU-UGCUUUUAUAGGCCAGACCAAAA---- ...((..((((...(((((.((((..((((((((((....)))))))(----(((((.((.....)).))))))..))))))-).)))))...))))...))....---- ( -33.30, z-score = -1.90, R) >dp4.chr4_group3 11290453 101 + 11692001 CCAGGAGGGCCAACAAAAGGGACUUGGUCAGCGACAUAGUUGUCGUCG----UUUGUAUGAGCUCCAAACGGGCGAGAUGGU-UGCUUUUAUAGGCCAGACCAAAA---- ...((..((((...(((((.((((..(((..(((((....)))))(((----(((((.((.....)).))))))))))))))-).)))))...))))...))....---- ( -34.10, z-score = -2.07, R) >droAna3.scaffold_12943 1205135 90 - 5039921 CCAGGAGGGCUAGCAGCAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAACCUUCGCCUGGUUCACCCAC-CGCUUUUAUAGGCCA------------ ...((.(((..(((..(((((....(((((((((((....))))))))-------......)))...))))))))..))).)-)((((....))))..------------ ( -31.80, z-score = -0.89, R) >droEre2.scaffold_4929 10381680 90 - 26641161 CCAGGAGGGCGAAUAGAAGGCACUUAGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAAGACCGGGUGAACCACU-UCCUUUUAUAGCCCU------------ .....(((((..((((((((((((..((((((((((....)))))))(-------((....)))..)))..)))).......-.)))))))).)))))------------ ( -36.00, z-score = -3.43, R) >droYak2.chr2L 12457791 90 - 22324452 CCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACCGGGCGAACCACU-UCCUUUUAUAGGCCC------------ ......((((..((((((((....((((..((((((....))))((((-------((....)))))).....))..))))..-.))))))))..))))------------ ( -35.30, z-score = -2.69, R) >droSec1.super_3 5216234 90 - 7220098 CCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCUUUAUAUAGGCCU------------ ......((((..(((.((((....(((..(((((((....))))((((-------((....)))))).....)))..)))..-.)))).)))..))))------------ ( -28.90, z-score = -1.07, R) >droSim1.chr2L 9547978 90 - 22036055 CUAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCCUUUUAUAGGCCU------------ ......((((..((((((((....(((..(((((((....))))((((-------((....)))))).....)))..)))..-.))))))))..))))------------ ( -35.20, z-score = -2.88, R) >consensus CCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG_______GUUUGAGCCAACACCGGGCGAGCCACU_UGCUUUUAUAGGCCA____________ ............................((((((((....)))))))).............................................................. ( -8.51 = -8.84 + 0.33)


| Location | 9,769,030 – 9,769,125 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.13 |
| Shannon entropy | 0.67434 |
| G+C content | 0.50656 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -9.45 |
| Energy contribution | -9.97 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
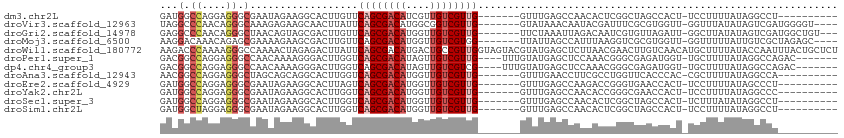
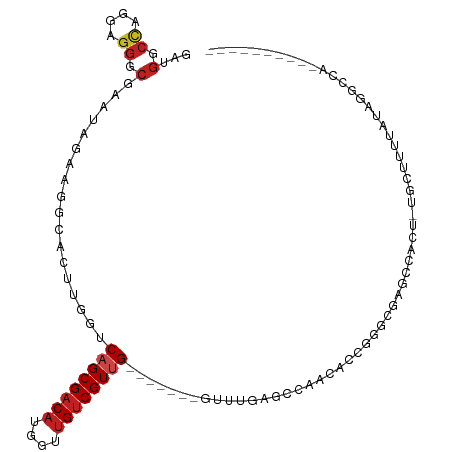
>dm3.chr2L 9769030 95 - 23011544 GAUGGCCAGGAGGGCGAAUAGAAGGCACUUGUUCAGCGACAUCGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCCUUUUAUAGGCCU---------- ...((((((((((.(.....)..(((......(((((((((....))))))))-------)....((((.......)))).))).))-))))......)))).---------- ( -34.40, z-score = -2.16, R) >droVir3.scaffold_12963 18358109 101 + 20206255 UAGGCCCAACAGGGCAAAGAGAAGCAACUUAUUCAGCGACAUGGCGGUCGUUG-------GUAUAAACAAUACGAUUUCGCGUGGUU-GGUUUAUAUAGUCGAUGGGGU---- ...((((....))))............(((((((((((((......)))))))-------((((((((..((((......))))...-.))))))))....))))))..---- ( -30.00, z-score = -2.44, R) >droGri2.scaffold_14978 474897 102 - 1124632 GAGGCCCAACAGGGCUAACAGUAGCGACUUGUUCAGCGACAUGGUUGUCGUUG-------UUCUAAAUUAGACAAUCGUGUUAGAUU-GGCUUAUAUAGUCGAUGGCUGU--- ..(((((....))))).(((((..(((((.(..((((((((....))))))))-------..).........(((((......))))-)........)))))...)))))--- ( -37.30, z-score = -3.71, R) >droMoj3.scaffold_6500 3544001 101 + 32352404 AAGGACAAACAGAGCGAAAAGAAGCGACUUGUUCAGCGACAUGGUUGUCGUGG-------UUAUUAGCCAUUUAAGGUCGCGUGGUU-GGUUUUUAUUGUCGCUAGAGC---- .(((((((..(((((.((.....(((((((.....((((((....))))))((-------(.....))).....)))))))....))-.)))))..))))).)).....---- ( -30.90, z-score = -2.13, R) >droWil1.scaffold_180772 1141548 113 + 8906247 AAGACCCAAAAGGGCCAAAACUAGAGACUUAUUCAGCGACAUGACUGCCGUUGGUAGUACGUAUGAGCUCUUAACGAACUUGUCAACAUGCUUUUAUACCAAUUUACUGCUCU ..(.(((....))).)......(((((((...((((((.((....)).)))))).)))..((((((((..((.(((....))).))...))))..))))..........)))) ( -18.60, z-score = 0.95, R) >droPer1.super_1 8405492 101 + 10282868 GACGGCCAGGAGGGCCAACAAAAGGGACUUGGUCAGCGACAUAGUUGUCGUUG----UUUGUAUGAGCUCCAAACGGGCGAGAUGGU-UGCUUUUAUAGGCCAGAC------- ...((((.((((((((((..........))))))(((((((....))))))).----..........))))....((((((.....)-))))).....))))....------- ( -33.00, z-score = -1.56, R) >dp4.chr4_group3 11290458 101 + 11692001 GACGGCCAGGAGGGCCAACAAAAGGGACUUGGUCAGCGACAUAGUUGUCGUCG----UUUGUAUGAGCUCCAAACGGGCGAGAUGGU-UGCUUUUAUAGGCCAGAC------- ...((((.((....))...(((((.((((..(((..(((((....)))))(((----(((((.((.....)).))))))))))))))-).)))))...))))....------- ( -33.50, z-score = -1.58, R) >droAna3.scaffold_12943 1205135 95 - 5039921 AACGGCCAGGAGGGCUAGCAGCAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAACCUUCGCCUGGUUCACCCAC-CGCUUUUAUAGGCCA---------- ...((((..((((((..((.....))....(((((((((((....))))))))-------(..((((((.......))))))..)))-)))))))...)))).---------- ( -37.80, z-score = -1.93, R) >droEre2.scaffold_4929 10381680 95 - 26641161 GAUGGCCAGGAGGGCGAAUAGAAGGCACUUAGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAAGACCGGGUGAACCACU-UCCUUUUAUAGCCCU---------- ..........(((((..((((((((((((..((((((((((....)))))))(-------((....)))..)))..)))).......-.)))))))).)))))---------- ( -36.00, z-score = -2.61, R) >droYak2.chr2L 12457791 95 - 22324452 GAUGGCCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACCGGGCGAACCACU-UCCUUUUAUAGGCCC---------- ...........((((..((((((((....((((..((((((....))))((((-------((....)))))).....))..))))..-.))))))))..))))---------- ( -35.30, z-score = -1.93, R) >droSec1.super_3 5216234 95 - 7220098 GAUGGCCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCUUUAUAUAGGCCU---------- ...((((((((((((........(((.....)))(((((((....))))((((-------((....)))))).....))).))).))-))).......)))).---------- ( -33.61, z-score = -1.70, R) >droSim1.chr2L 9547978 95 - 22036055 GAUGGCUAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG-------GUUUGAGCCAACACUCGGCUAGCCACU-UCCUUUUAUAGGCCU---------- ...........((((..((((((((....(((..(((((((....))))((((-------((....)))))).....)))..)))..-.))))))))..))))---------- ( -35.20, z-score = -2.06, R) >consensus GAUGGCCAGGAGGGCGAAUAGAAGGCACUUGGUCAGCGACAUGGUUGUCGUUG_______GUUUGAGCCAACACCGGGCGAGCCACU_UGCUUUUAUAGGCCA__________ ...(.((....)).)..................((((((((....))))))))............................................................ ( -9.45 = -9.97 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:55 2011