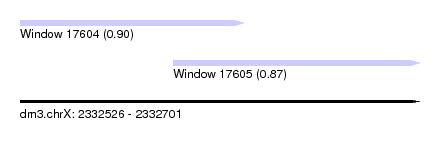
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,332,526 – 2,332,701 |
| Length | 175 |
| Max. P | 0.895525 |

| Location | 2,332,526 – 2,332,624 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Shannon entropy | 0.28894 |
| G+C content | 0.59008 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
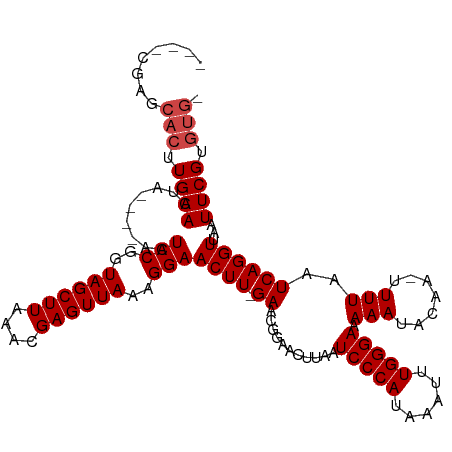
>dm3.chrX 2332526 98 + 22422827 GGCCUGAGUGGCCUGAGAGCCCGGGGCGGAUGGCCGAAUGGCCUGAGAUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA ((((.....))))...(((((((((((.....))).....(((((.(((((((...)))))))))))).................))))..))))... ( -42.40, z-score = -1.73, R) >droSim1.chrX 1650728 89 + 17042790 ---------GACCUGAGAGCCCUGGGCGGAUGGCCGAGUGGCCUGAGAUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA ---------(.((((.((.(((((((.((((((((....))))....)))))))))))...)))))))(((((((.((......)).))).))))... ( -37.00, z-score = -2.01, R) >droSec1.super_10 2105035 89 + 3036183 ---------GACCUGAGAGCCCUGGGCGGAUGGCCGAGUGGCCUGAGAUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA ---------(.((((.((.(((((((.((((((((....))))....)))))))))))...)))))))(((((((.((......)).))).))))... ( -37.00, z-score = -2.01, R) >droYak2.chrX 15755679 88 - 21770863 -------GAUGCCUGGGUUGCCUGAGUGGC---CUGGGAUGCCUGAGAUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA -------....((..(((..(....)..))---)..))..(((((.(((((((...))))))))))))(((((((.((......)).))).))))... ( -40.10, z-score = -3.40, R) >droEre2.scaffold_4644 2280359 95 + 2521924 GGCCUGUGUAGCCUGAGUGUCCCGAGUGUC---CUGAGUGGCCUGAGGUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA (((.......)))(((((...(((..(...---..((((((((((.(((((((...))))))))))))...))))).......)..)))..))))).. ( -33.00, z-score = -0.77, R) >consensus _______G_GGCCUGAGAGCCCUGGGCGGAUGGCCGAGUGGCCUGAGAUCUCCCGGGGAGAUCCAGGCGAGCACUUGAACUAAAUCCGGUAGCUUAAA .....................((.(((.....))).))..(((((.(((((((...))))))))))))(((((((.((......)).))).))))... (-27.92 = -28.20 + 0.28)
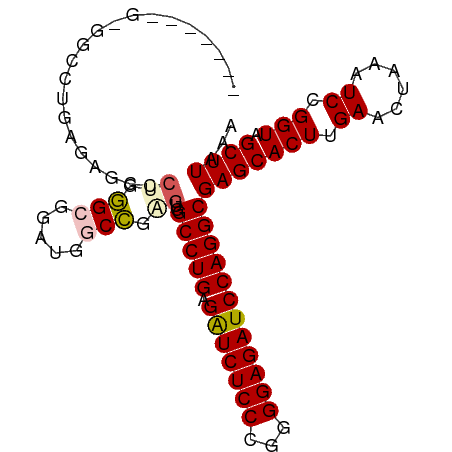
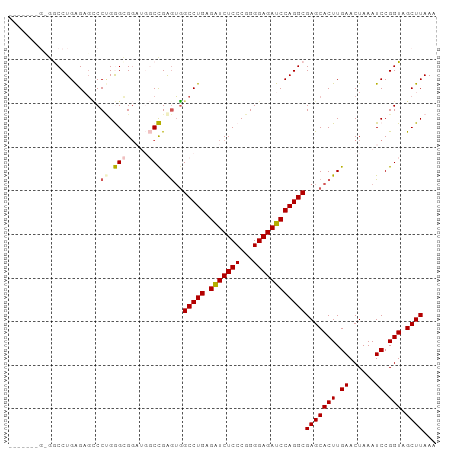
| Location | 2,332,593 – 2,332,701 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Shannon entropy | 0.12027 |
| G+C content | 0.34977 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2332593 108 + 22422827 ----CGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----...((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))).- ( -24.00, z-score = -1.96, R) >droSim1.chrX 1650786 108 + 17042790 ----CGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----...((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))).- ( -24.00, z-score = -1.96, R) >droSec1.super_10 2105093 108 + 3036183 ----CGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----...((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))).- ( -24.00, z-score = -1.96, R) >droYak2.chrX 15755736 108 - 21770863 ----CGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----...((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))).- ( -24.00, z-score = -1.96, R) >droEre2.scaffold_4644 2280423 108 + 2521924 ----CGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----...((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))).- ( -24.00, z-score = -1.96, R) >droAna3.scaffold_12929 2238036 109 + 3277472 ---CGAGGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ---((((..((((((....----..(((..((((((....))))))..)))....-.............(((((......))))).........-......))))))...))))....- ( -25.20, z-score = -2.08, R) >dp4.chrXL_group1a 3506312 111 + 9151740 -GAUUGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- -..(((((((((.((....----..)).))).))))))((((((((.....((((-((...........(((((......)))))(((((...)-))))..))))))))))))))...- ( -25.70, z-score = -2.40, R) >droPer1.super_11 46634 111 + 2846995 -GAUUGAGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- -..(((((((((.((....----..)).))).))))))((((((((.....((((-((...........(((((......)))))(((((...)-))))..))))))))))))))...- ( -25.70, z-score = -2.40, R) >droWil1.scaffold_181150 2040013 114 - 4952429 ----AGAGCACUUGAACUAUGUAAAUCCGGUAGCUUAAACGAGUUAAAGGAACUUUGAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGCAGAG ----.(((.((((((....((((..(((..((((((....))))))..)))..................(((((......)))))....)))).-......))))))...)))...... ( -22.70, z-score = -1.31, R) >droVir3.scaffold_12928 296138 112 - 7717345 AGGUCGCGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ......(((((........----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))......)))))- ( -25.10, z-score = -1.49, R) >droMoj3.scaffold_6473 2628289 102 + 16943266 ----------CUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA-UUUUAAUCAGGUUAAUUCGUGUG- ----------...(((...----..(((..((((((....))))))..)))((((-((...........(((((......)))))(((((...)-))))..))))))...))).....- ( -20.50, z-score = -1.37, R) >droGri2.scaffold_15081 3284536 113 + 4274704 AGGUCGCGCACUUGAACUA----AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU-GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAAAUUUUAAUCAGGUUAAUUCGUUUG- .....((..(((.((....----..)).))).)).(((((((((((.....((((-((...........(((((......)))))(((((....)))))..)))))))))))))))))- ( -24.70, z-score = -1.51, R) >consensus ____CGAGCACUUGAACUA____AAUCCGGUAGCUUAAACGAGUUAAAGGAACUU_GAACGGAACUUAAUCCCAUAAAUUUGGGAAAAAUACAA_UUUUAAUCAGGUUAAUUCGUGUG_ .........((((((..........(((..((((((....))))))..)))..................(((((......)))))................))))))............ (-21.40 = -21.48 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:09 2011