| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,273,191 – 2,273,265 |
| Length | 74 |
| Max. P | 0.500000 |

| Location | 2,273,191 – 2,273,265 |
|---|---|
| Length | 74 |
| Sequences | 13 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.68797 |
| G+C content | 0.56783 |
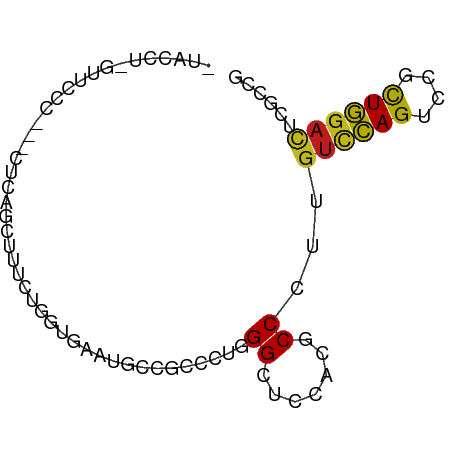
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -7.59 |
| Energy contribution | -7.25 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
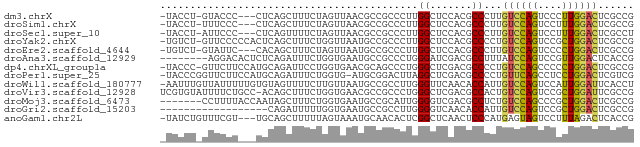
>dm3.chrX 2273191 74 - 22422827 -UACCU-GUACCC---CUCAGCUUUCUAGUUAACGCCGCCCUUGGCUCCACGCCCUUGUCCAGUCCCUUGGACUCGCCG -.....-......---...((((....))))...((((....)))).....((....((((((....))))))..)).. ( -12.60, z-score = -0.36, R) >droSim1.chrX 1595775 74 - 17042790 -UACCU-UUUCCC---CUCAGCUUUCUAGUUAACGCCGCCCUUGGCUCCACGCCCUUGUCCAGUCCUUUGGACUCGCCG -.....-......---...((((....))))...((((....)))).....((....((((((....))))))..)).. ( -13.40, z-score = -1.38, R) >droSec1.super_10 2053883 74 - 3036183 -UACCU-AUUCCC---CUCAGUUUUCUAGUUAACGCCGCCCUUGGCUCCACGCCCUUGUCCAGUCCUUUGGACUCGCCU -.....-......---....(((........)))((((....)))).....((....((((((....))))))..)).. ( -12.70, z-score = -1.19, R) >droYak2.chrX 15707443 77 + 21770863 -UGUCU-GUUCCCCCACUCAGCUUUCUGGUUAAUGCCGCCCUUGGCUCCACGCCCCUGUCCAGUCCGCUGGACUCGCCG -.....-.............((.....(((....((((....)))).....)))...((((((....))))))..)).. ( -17.60, z-score = -0.83, R) >droEre2.scaffold_4644 2233525 74 - 2521924 -UGUCU-GUAUUC---CACAGCUUUCUAGUUAAUGCCGCCCUUGGCUCCACGCCCUUGUCCAGUCCCCUGGACUCGCCG -.(.((-((....---.)))))......((....((((....))))...))((....((((((....))))))..)).. ( -15.70, z-score = -1.43, R) >droAna3.scaffold_12929 2182630 71 - 3277472 --------AGGACACUCUCAGAUUUCUGGUGAAUGCCGCCCUGGGAUCGACGCCUUUAUCCAGUCCGUUGGACUCACCG --------.((.........(((..(.((((.....))))..)..))).............(((((...)))))..)). ( -17.10, z-score = 0.53, R) >dp4.chrXL_group1a 6097949 77 - 9151740 -UACCC-GUUCUUCCAUGCAGAUUCCUGGUGAACGCAGCCCUGGGCUCGACGCCCCUGUCCAGCCCCCUGGACUCGCCG -.....-((((..(((.(......).))).))))((((.((.(((((.((((....)))).)))))...)).)).)).. ( -23.10, z-score = -1.15, R) >droPer1.super_25 175857 77 + 1448063 -UACCCGGUUCUUCCAUGCAGAUUUCUGGUG-AUGCGGACUUAGGCUCGACGCCCCUGUUCAGCCUCCUGGACUCGUCG -...((((.(((.......)))...)))).(-(((.(..(..(((((.((((....)))).)))))...)..).)))). ( -20.30, z-score = 0.24, R) >droWil1.scaffold_180777 1976078 78 - 4753960 -AAUUUGUUAUUUUUGUGUAGUUUUCUUGUUAAUGCCGCCUUGGGUUCAACACCAUUGUCCAGUCCAUUGGAUUCACCU -..............(((.........((((...(((......)))..)))).....((((((....)))))).))).. ( -9.90, z-score = 0.89, R) >droVir3.scaffold_12928 5316590 78 + 7717345 UCGUGUAUUUUCUGCC-ACAGCUUUCUGGUGAACGCCGCCCUGGGCUCGACGCCACUGUCCAGUCCGCUGGAUUCGCCG ..(((..........)-))........((((((..((((...(((((.((((....)))).))))))).)).)))))). ( -21.60, z-score = 0.16, R) >droMoj3.scaffold_6473 2771691 72 + 16943266 -------CCUUUUACCAAUAGCUUUCUGGUGAAUGCCGCAUUGGGGUCGACGCCUCUGUCCAGCCCGCUGGACUCGCCG -------...(((((((.........)))))))....((...(((((....))))).((((((....))))))..)).. ( -22.30, z-score = -1.04, R) >droGri2.scaffold_15203 2451112 61 + 11997470 ------------------CAGAUUUUUGGUGAAUGCCGCCUUGGGGUCAACACCAUUGUCCAGUCCGCUGGACUCGCCG ------------------........(((((...(((.(....))))...)))))..((((((....))))))...... ( -19.80, z-score = -1.17, R) >anoGam1.chr2L 47194193 75 + 48795086 -UAUCUGUUUCGU---UGCAGCUUUUUAGUAAAUGCAACACUCGGCUCAACUCCCAUGAGUAGUCCUUUAGACUCACCG -..((((....((---(((((((....)))...))))))....((((..((((....))))))))...))))....... ( -15.10, z-score = -1.71, R) >consensus _UACCU_GUUCCC___CUCAGCUUUCUGGUGAAUGCCGCCCUGGGCUCCACGCCCUUGUCCAGUCCGCUGGACUCGCCG ...........................................((.......))...((((((....))))))...... ( -7.59 = -7.25 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:04 2011