
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,268,114 – 2,268,234 |
| Length | 120 |
| Max. P | 0.511355 |

| Location | 2,268,114 – 2,268,234 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Shannon entropy | 0.31289 |
| G+C content | 0.59306 |
| Mean single sequence MFE | -46.71 |
| Consensus MFE | -20.53 |
| Energy contribution | -19.31 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2268114 120 - 22422827 CAAAGUGGUGGCCAGUAGCAGUCGUCGGGAAACCGGCGACACCGAGAGCGACGUGGCCCAACUGAUAACCGAUGAAAUUUCCCAAUCGCAAUCAGAGGCCACCGAUGACAGCGUCGGAGC .....(((((((((((....((((((((....)))))))).((....).))).)))))...(((((...((((...........))))..)))))..))))((((((....))))))... ( -46.80, z-score = -3.25, R) >droGri2.scaffold_15203 2444140 120 + 11997470 CAAGGUAGUCGCUAGCAGUAGUCGGCGAGAAACGGGCGACACCGAAAGCGAUGUGGCCCAACUAAUCACCGAUGAACUGUCGCUGUCGCAAUCAGAGGCAACCGAUGACAGCGUCGGCGC ..............(((((.(((((.((.....((((.(((((....).).))).))))......)).)))))..)))))(((((.(((..(((..((...))..)))..))).))))). ( -45.50, z-score = -2.01, R) >droMoj3.scaffold_6473 2764041 120 + 16943266 GAAGGUAGUCGCUAGCAGCAGCCGGCGCGAGACCGGCGACACGGAGAGCGAUGUGGCCCAACUAAUCACCGACGAACUGUCGCUGUCCCAAUCAGAGGCGACAGAUGACAGCGUCAACGC ...(((.((((((..(.(..((((((....).)))))..)...)..))))))...)))............((((..(((((((((((((.......)).))))).))))))))))..... ( -49.10, z-score = -2.85, R) >droVir3.scaffold_12928 5309836 120 + 7717345 GAAGGUAGUUGCUAGCAGCAGCCGGCGCGAGACCGGCGACACCGAAAGCGAUGUGGCCCAACUAAUCACCGAUGAACUCUCGCUGUCCCAAUCAGAGGCCACCGAUGACAGCGUCGGCGC ...(((.((((....)))).((((((....).)))))...)))....(((..((((((...........(((.(....))))(((.......))).))))))(((((....))))).))) ( -40.70, z-score = -0.45, R) >droWil1.scaffold_180777 1968806 120 - 4753960 CAAGGUAGUGGCUAGCAGUAGUCGUCGCGAAACCGGCGACACCGAAAGCGAUGUGGCCCAAUUGAUCACCGAUGAAUUGUCCUUAUCGCAAUCAGAGGCCACCGAUGAUAGUGUCGGCGC ....((.((((((.......(((((((......)))))))...((..((((((.((..(((((.(((...))).))))).)).))))))..))...))))))((((......)))))).. ( -39.80, z-score = -1.26, R) >droPer1.super_25 169981 120 + 1448063 GAAGGUAGUCGCUAGCAGCAGUCGCCGGGAGACCGGCGAUACGGAGAGCGAUGUAGCCCAGCUGAUCACCGACGAACUGUCCCUCUCCCAAUCAGAGGCCACGGACGAGAGCGUUGGCGC ...(((.((((((..(....((((((((....))))))))...)..))))))...)))..........((((((..((((((.((((.......))))....)))).))..))))))... ( -48.70, z-score = -1.82, R) >dp4.chrXL_group1a 6092357 120 - 9151740 GAAGGUAGUCGCUAGCAGCAGUCGCCGGGAGACCGGCGAUACGGAGAGCGAUGUAGCCCAGCUGAUCACCGACGAACUGUCCCUCUCCCAAUCAGAGGCCACGGACGAGAGCGUUGGCGC ...(((.((((((..(....((((((((....))))))))...)..))))))...)))..........((((((..((((((.((((.......))))....)))).))..))))))... ( -48.70, z-score = -1.82, R) >droAna3.scaffold_12929 2177167 120 - 3277472 CAAAGUAGUGGCUAGCAGCAGUCGCCGGGAGACCGGCGACACGGAGAGCGAUGUCGCCCAACUGAUCACCGACGAAAUCUCGAUCUCCCAAUCAGAGGCGACGGAUGACAGUGUUGGGGC .......((.(((..(....((((((((....))))))))...)..))).))...((((.((((.((((((.((...(((.(((......))))))..)).))).)))))))....)))) ( -50.10, z-score = -2.95, R) >droEre2.scaffold_4644 2228278 120 - 2521924 CAAGGUGGUGGCCAGUAGCAGUCGUCGGGAAACCGGCGACACAGAGAGCGAUGUCGCCCAACUGAUAACCGAUGAAAUUUCCCAAUCGCAAUCAGAGGCCACCGAUGACAGUGUCGGGGC ......(((((((.......((((((((....)))))))).....(.(((....))).)..(((((...((((...........))))..))))).)))))))(((......)))..... ( -46.60, z-score = -2.86, R) >droYak2.chrX 15701884 120 + 21770863 CAAGGUGGUGGCCAGUAGCAGUCGUCGGGAAACCGGCGACACGGAGAGCGACGUGGCCCAACUGAUAACCGAUGAAAUUUCCCAAUCGCAAUCAGAGGCCACCGAUGACAGCGUCGGUGC ...((((((((((.......((((((((....))))))))(((........)))))))...(((((...((((...........))))..)))))..))))))((((....))))..... ( -49.40, z-score = -3.11, R) >droSec1.super_10 2048938 120 - 3036183 CAAAGUGGUGGCCAGUAGCAGUCGUCGGGAAACCGGCGACACCGAGAGUGACGUGGCUCAACUGAUAACCGAUGAAAUUUCCCAAUCGCAAUCAGAGGCCACCGAUGAUAGCGUCGGAGC ......(((((((.......((((((((....)))))))).....((((......))))..(((((...((((...........))))..))))).)))))))((((....))))..... ( -48.30, z-score = -3.83, R) >droSim1.chrX 1590722 120 - 17042790 CAAAGUGGUGGCCAGUAGCAGUCGUCGGGAAACCGGCGACACCGAGAGUGACGUGGCCCAACUGAUAACCGAUGAAAUUUCCCAAUCGCAAUCAGAGGCCACCGAUGACAGCGUCGGAGC .....(((((((((((....((((((((....)))))))).((....).))).)))))...(((((...((((...........))))..)))))..))))((((((....))))))... ( -46.80, z-score = -3.38, R) >consensus CAAGGUAGUGGCUAGCAGCAGUCGUCGGGAAACCGGCGACACCGAGAGCGAUGUGGCCCAACUGAUCACCGAUGAAAUGUCCCUAUCGCAAUCAGAGGCCACCGAUGACAGCGUCGGCGC .....((((.((((...((.(((((((......))))))).......))....))))...))))....((((((.....((...((((..............))))))...))))))... (-20.53 = -19.31 + -1.22)
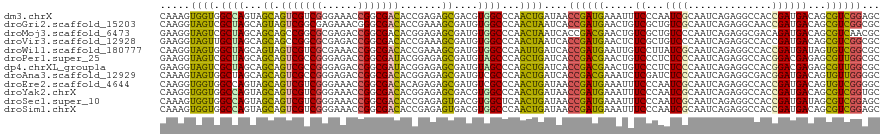

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:03 2011