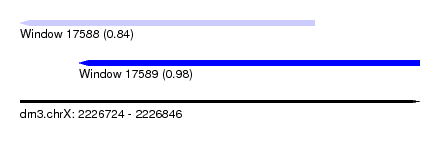
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,226,724 – 2,226,846 |
| Length | 122 |
| Max. P | 0.980872 |

| Location | 2,226,724 – 2,226,814 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.41274 |
| G+C content | 0.36016 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -9.12 |
| Energy contribution | -11.23 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2226724 90 - 22422827 ACAUUGAACCACAUUACUAUAUAUGGAGUGUGGUUGAAAACCGAGUUUCCAAAGGAGAGUCAUAAAAGCAAGCAAUAUCAAAAACAAAAA ...((.(((((((((.(((....)))))))))))).))....((.(((((...))))).))............................. ( -19.80, z-score = -3.05, R) >droSec1.super_10 2003104 84 - 3036183 ACAUUGAACCACAUUACUAUAUAUAGCGUGUGGUUGGAAACCGAGUUUCCAAAGGAGAGUCAU-AAAGCAAGCAAUAUCAAAAGC----- ...((.((((((((..(((....))).)))))))).))....((.(((((...))))).))..-.....................----- ( -18.00, z-score = -2.16, R) >droAna3.scaffold_12929 2126279 71 - 3277472 ACAUUGAACCACAUUACUAUAUAUAGAGUAUGGAGGGACUC-----------------GUCGGAAAAACAAACAAUGACAAACUCAGA-- .(((((..((.(((.(((........))))))..))((...-----------------.))...........)))))...........-- ( -9.10, z-score = -0.24, R) >consensus ACAUUGAACCACAUUACUAUAUAUAGAGUGUGGUUGGAAACCGAGUUUCCAAAGGAGAGUCAUAAAAGCAAGCAAUAUCAAAAACA_A__ ..(((((((((((((.(((....)))))))))))).......((.((((.....)))).))...........)))).............. ( -9.12 = -11.23 + 2.11)


| Location | 2,226,742 – 2,226,846 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Shannon entropy | 0.08663 |
| G+C content | 0.39749 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -21.41 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
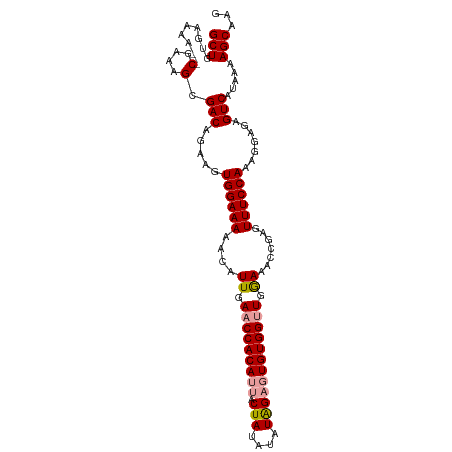
>dm3.chrX 2226742 104 - 22422827 GCUGUGAAAA--CGAAAGCGACAGAAGUGGAAAAACAUUGAACCACAUUACUAUAUAUGGAGUGUGGUUGAAAACCGAGUUUCCAAAGGAGAGUCAUAAAAGCAAG (((((....)--)......(((.....((((((....((.(((((((((.(((....)))))))))))).)).......)))))).......))).....)))... ( -25.30, z-score = -3.13, R) >droSec1.super_10 2003117 103 - 3036183 GCUGUGAAAA--CGAAAGCGACAGAAGUGGAAAAACAUUGAACCACAUUACUAUAUAUAGCGUGUGGUUGGAAACCGAGUUUCCAAAGGAGAGUCAU-AAAGCAAG (((((....)--)......(((...((((......)))).((((((((..(((....))).))))))))((((((...))))))........)))..-..)))... ( -24.70, z-score = -2.55, R) >droEre2.scaffold_4644 2186702 105 - 2521924 GCUGUGAAAAACCGAAAGCGACAGAAGUGGAAAAACAUUGAACCACA-UACUAUAUAUAGAGUGUGGCUGGAAACCGAGUUUCCAAAGUAGAGUCAUAAAAGCAAA (((.........(....).(((...((((......))))...(((((-(.(((....))).)))))).(((((((...))))))).......))).....)))... ( -24.00, z-score = -2.85, R) >consensus GCUGUGAAAA__CGAAAGCGACAGAAGUGGAAAAACAUUGAACCACAUUACUAUAUAUAGAGUGUGGUUGGAAACCGAGUUUCCAAAGGAGAGUCAUAAAAGCAAG (((.........(....).(((.....((((((....((.(((((((((.(((....)))))))))))).)).......)))))).......))).....)))... (-21.41 = -21.97 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:56 2011