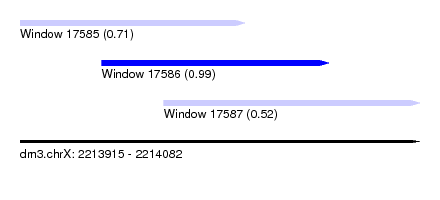
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,213,915 – 2,214,082 |
| Length | 167 |
| Max. P | 0.987987 |

| Location | 2,213,915 – 2,214,009 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.38 |
| Shannon entropy | 0.22383 |
| G+C content | 0.61918 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -27.66 |
| Energy contribution | -28.35 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.710309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2213915 94 + 22422827 UUCCCAUCCCCCUUACCCCGGGCCAGGAUUUUCCCAUUUCGACUGACAGGC-----GGUGGCGGCGGUGGACGUCAGCGUUUCUGUUGACGUCUCCCGU ..((((((((..(((...((((...((.....))...))))..)))..)).-----))))).)((((..((((((((((....))))))))))..)))) ( -33.80, z-score = -0.60, R) >droEre2.scaffold_4644 2171411 92 + 2521924 -------UUCCCACCCUCUGUGCCCGGAUUUUCCCAUUUCGACUGACAGGCAGAGUGGCGGUGGCGGUGGACGUCAUCGUUUCUGUUGACGUCUCCCGU -------...(((((((((.((((.((......))...((....))..)))).)).)).)))))(((..(((((((.((....)).)))))))..))). ( -35.00, z-score = -1.60, R) >droSec1.super_10 1990715 88 + 3036183 ------UUCCCACCCCCCUGUGCCCGGAUUUUCCCAUUUCGACUGACAGGC-----GGUGGCGGCGGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGU ------..((((((..(((((...((((.........))))....))))).-----))))).)((((.(((((((((((....))))))))))).)))) ( -37.50, z-score = -2.49, R) >droSim1.chrX 1561412 88 + 17042790 ------UUCCCACCCCCCUGUGCCCGGAUUUUCCCAUUUCGACUGACAGGC-----GGUGGCGGCGGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGU ------..((((((..(((((...((((.........))))....))))).-----))))).)((((.(((((((((((....))))))))))).)))) ( -37.50, z-score = -2.49, R) >consensus ______UUCCCACCCCCCUGUGCCCGGAUUUUCCCAUUUCGACUGACAGGC_____GGUGGCGGCGGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGU .........(((((.......(((.((......))...((....))..))).....)))))..((((..((((((((((....))))))))))..)))) (-27.66 = -28.35 + 0.69)

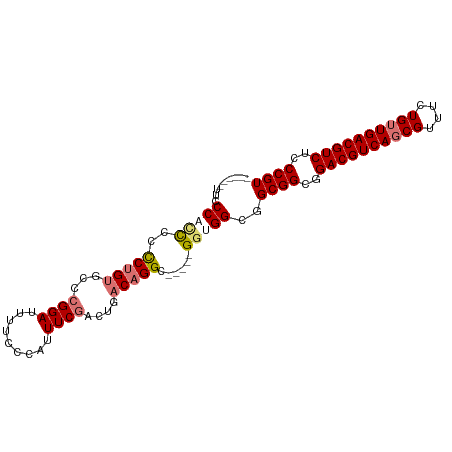
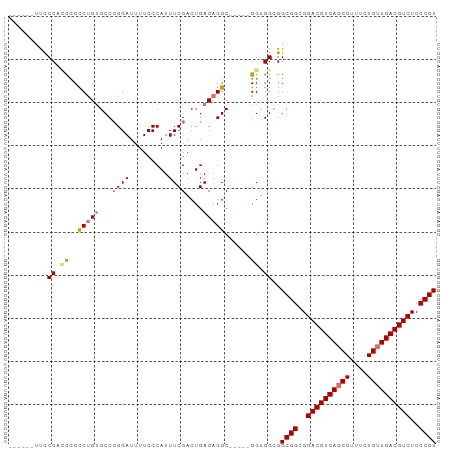
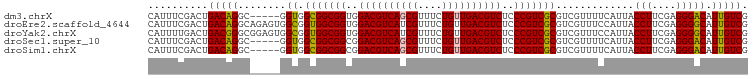
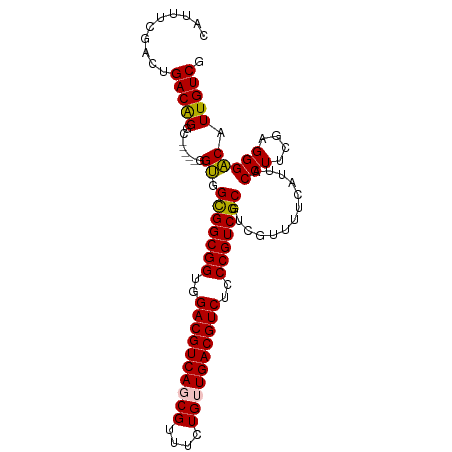
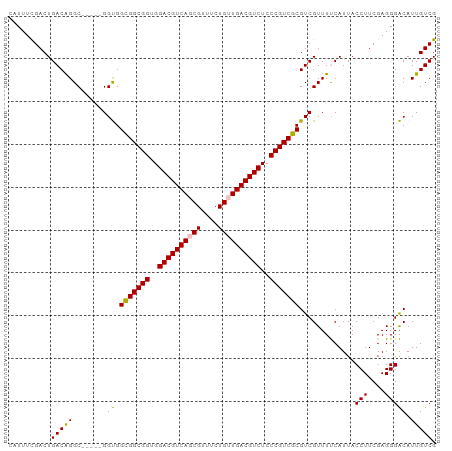
| Location | 2,213,949 – 2,214,044 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Shannon entropy | 0.12524 |
| G+C content | 0.58558 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -37.84 |
| Energy contribution | -37.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2213949 95 + 22422827 CAUUUCGACUGACAGGC-----GGUGGCGGCGGUGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCG ..............(((-----(((((((((((..((((((((((....))))))))))..))))))).............(((....))).))))))). ( -42.20, z-score = -3.39, R) >droEre2.scaffold_4644 2171438 100 + 2521924 CAUUUCGACUGACAGGCAGAGUGGCGGUGGCGGUGGACGUCAUCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUCCAUUACCUUCGAGGGGCAUUGUCG ..........(((((((.((((((((.((((((..(((((((.((....)).)))))))..))))))))))))))......(((....))))).))))). ( -38.50, z-score = -1.58, R) >droYak2.chrX 15649714 100 - 21770863 CAUUUUGACUGACGGGCGGAGUGGCGGUGGCGGUGGACGUCAUCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUCCAUUACCUUCGAGGGGCAUUGUCG ..........((((((((((..((((.((((((..(((((((.((....)).)))))))..))))))))))...)))....(((....))))).))))). ( -40.50, z-score = -1.72, R) >droSec1.super_10 1990743 95 + 3036183 CAUUUCGACUGACAGGC-----GGUGGCGGCGGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCG ..............(((-----(((((((((((.(((((((((((....))))))))))).))))))).............(((....))).))))))). ( -41.60, z-score = -3.03, R) >droSim1.chrX 1561440 95 + 17042790 CAUUUCGACUGACAGGC-----GGUGGCGGCGGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCG ..............(((-----(((((((((((.(((((((((((....))))))))))).))))))).............(((....))).))))))). ( -41.60, z-score = -3.03, R) >consensus CAUUUCGACUGACAGGC_____GGUGGCGGCGGUGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCG .....((((.(((..((......)).(((((((..((((((((((....))))))))))..))))))))))..........(((....))).....)))) (-37.84 = -37.60 + -0.24)
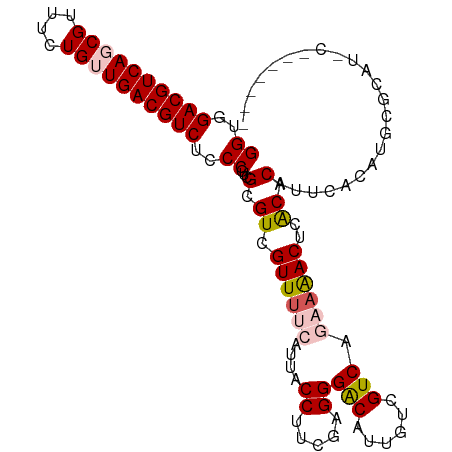
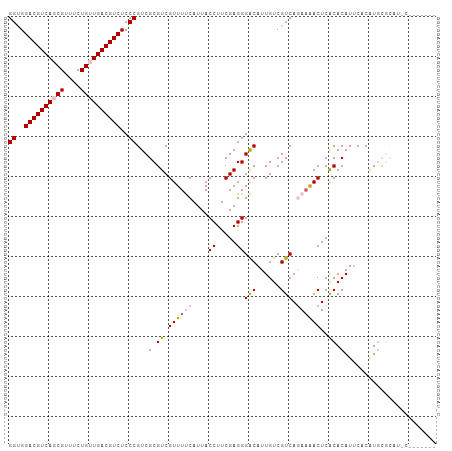
| Location | 2,213,975 – 2,214,082 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Shannon entropy | 0.22296 |
| G+C content | 0.52349 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2213975 107 + 22422827 GGUGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCGUCACAAGACUCACACAUUCACAUUCACAUACAACAUGC ((..((((((((((....))))))))))..))(((.(.(((.............))).))))..(((.(((....)))..)))........................ ( -28.42, z-score = -1.38, R) >droEre2.scaffold_4644 2171469 100 + 2521924 GGUGGACGUCAUCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUCCAUUACCUUCGAGGGGCAUUGUCGUCACACAACUCGCACAUUCUCGUGUGCAUAC------- ((..(((((((.((....)).)))))))..))((.(((.((...((....((.....))))...)).))).)).......((((((....))))))....------- ( -29.70, z-score = -1.22, R) >droYak2.chrX 15649745 100 - 21770863 GGUGGACGUCAUCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUCCAUUACCUUCGAGGGGCAUUGUCGUCAGAAAACUCGCACAUACUCGUGUGCAUGC------- ((((.....)))).((((((.(((((...(((.(((....((.......))...))).)))...))))).))))))....((((((....))))))....------- ( -30.20, z-score = -0.75, R) >droSec1.super_10 1990769 94 + 3036183 GGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCGUCAGAAAACUCACACAUUCACAUGC------------- ((.(((((((((((....))))))))))).))...(.((.((((((....((.....))(((......))).))))))..)).)..........------------- ( -29.00, z-score = -1.76, R) >droSim1.chrX 1561466 96 + 17042790 GGCGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCGUCAGAAAACUCACACAUUCACAUGCGU----------- ((.(((((((((((....))))))))))).))..(((((.((((((....((.....))(((......))).))))))............))))).----------- ( -29.92, z-score = -1.64, R) >consensus GGUGGACGUCAGCGUUUCUGUUGACGUCUCCCGUCGCGUCGUUUUCAUUACCUUCGAGGGACAUUGUCGUCAGAAAACUCACACAUUCACAUGCGCAU_C_______ ((..((((((((((....))))))))))..))...(.((.((((((....((.....))(((......))).))))))..)).)....................... (-24.16 = -24.92 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:54 2011