| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,196,413 – 2,196,513 |
| Length | 100 |
| Max. P | 0.961551 |

| Location | 2,196,413 – 2,196,513 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Shannon entropy | 0.26687 |
| G+C content | 0.49365 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.78 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
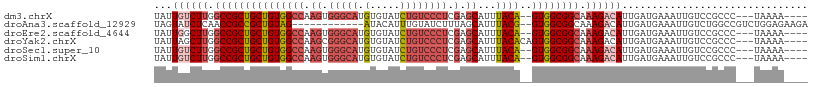
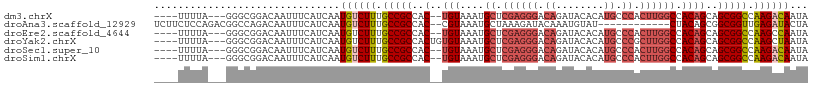
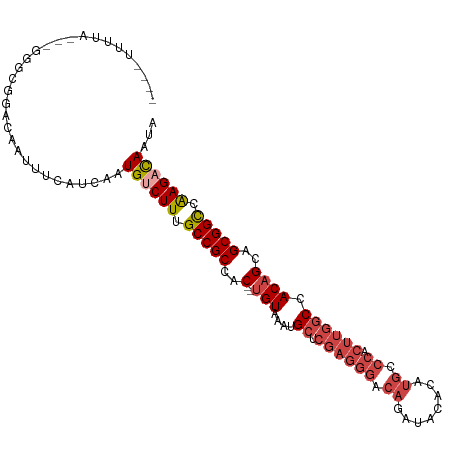
>dm3.chrX 2196413 100 + 22422827 UAUUGUCUUGGCCGCUGCUGUGGCCAAGUGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACA--GUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC---UAAAA---- ...((((((.(((((..((((((((.((.(((((........))))))).).))...))))--)..))))).))))))...................---.....---- ( -37.00, z-score = -2.73, R) >droAna3.scaffold_12929 2102730 95 + 3277472 UAGUAUCUCAACCGCCGCUGUAG------------AUACAUUUGUAUCUUUAGCAUUUACG--GUGGCGGCAAAGACAUUGAUGAAAUUGUCUGGCCGUCUGGAGAAGA .....((((.((((..((((.((------------((((....)))))).)))).....))--))((((((..(((((.(......).))))).))))))..))))... ( -34.80, z-score = -3.91, R) >droEre2.scaffold_4644 2162318 100 + 2521924 UAUUGGCUUGGCCGCUGCUGUGGCCAAGUGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACA--GUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC---UAAAA---- ....(((...(((((..((((((((.((.(((((........))))))).).))...))))--)..)))))...((((.(......).)))).))).---.....---- ( -36.70, z-score = -1.98, R) >droYak2.chrX 15640212 102 - 21770863 UAUUAGCUUGGCCGCUGCUGUGGCCAAGCGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACACAGUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC---UAAAA---- .....((((((((((....))))))))))((((.((((((..(((.......)))..))))))...........((((.(......).)))).))))---.....---- ( -40.10, z-score = -3.01, R) >droSec1.super_10 1981655 100 + 3036183 UAUUGUCUUGGCCGCUGCUGUGGCCAAGUGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACA--GUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC---UAAAA---- ...((((((.(((((..((((((((.((.(((((........))))))).).))...))))--)..))))).))))))...................---.....---- ( -37.00, z-score = -2.73, R) >droSim1.chrX 1552366 100 + 17042790 UAUUGUCUUGGCCGCUGCUGUGGCCAAGUGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACA--GUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC---UAAAA---- ...((((((.(((((..((((((((.((.(((((........))))))).).))...))))--)..))))).))))))...................---.....---- ( -37.00, z-score = -2.73, R) >consensus UAUUGUCUUGGCCGCUGCUGUGGCCAAGUGGGCAUGUGUAUCUGUCCCUCGAGCAUUUACA__GUGGCGGCAAAGACAUUGAUGAAAUUGUCCGCCC___UAAAA____ ...((((((.((((((((.(((.((....)).))).((((..(((.......)))..))))..)))))))).))))))............................... (-23.14 = -24.78 + 1.64)
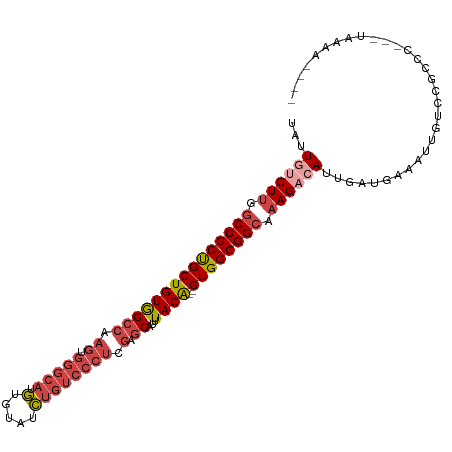
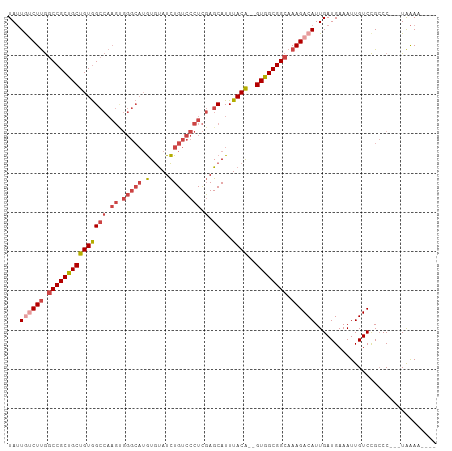
| Location | 2,196,413 – 2,196,513 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Shannon entropy | 0.26687 |
| G+C content | 0.49365 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -21.31 |
| Energy contribution | -23.17 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2196413 100 - 22422827 ----UUUUA---GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCAC--UGUAAAUGCUCGAGGGACAGAUACACAUGCCCACUUGGCCACAGCAGCGGCCAAGACAAUA ----...((---((((((.(((...((....))...)))))))).)--))...........(((.((........))))).(((((((.(....).)))))))...... ( -32.10, z-score = -2.45, R) >droAna3.scaffold_12929 2102730 95 - 3277472 UCUUCUCCAGACGGCCAGACAAUUUCAUCAAUGUCUUUGCCGCCAC--CGUAAAUGCUAAAGAUACAAAUGUAU------------CUACAGCGGCGGUUGAGAUACUA ...((((..(.((((.(((((..........)))))..)))).)((--(((...((((..((((((....))))------------))..))))))))).))))..... ( -29.10, z-score = -3.70, R) >droEre2.scaffold_4644 2162318 100 - 2521924 ----UUUUA---GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCAC--UGUAAAUGCUCGAGGGACAGAUACACAUGCCCACUUGGCCACAGCAGCGGCCAAGCCAAUA ----...((---((((((.(((...((....))...)))))))).)--))...........(((.((........))))).(((((((.(....).)))))))...... ( -31.40, z-score = -1.75, R) >droYak2.chrX 15640212 102 + 21770863 ----UUUUA---GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCACUGUGUAAAUGCUCGAGGGACAGAUACACAUGCCCGCUUGGCCACAGCAGCGGCCAAGCUAAUA ----.....---((((((.(((...((....))...))))).....((((((..((((....)).))..)))))).))))((((((((.(....).))))))))..... ( -37.20, z-score = -2.69, R) >droSec1.super_10 1981655 100 - 3036183 ----UUUUA---GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCAC--UGUAAAUGCUCGAGGGACAGAUACACAUGCCCACUUGGCCACAGCAGCGGCCAAGACAAUA ----...((---((((((.(((...((....))...)))))))).)--))...........(((.((........))))).(((((((.(....).)))))))...... ( -32.10, z-score = -2.45, R) >droSim1.chrX 1552366 100 - 17042790 ----UUUUA---GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCAC--UGUAAAUGCUCGAGGGACAGAUACACAUGCCCACUUGGCCACAGCAGCGGCCAAGACAAUA ----...((---((((((.(((...((....))...)))))))).)--))...........(((.((........))))).(((((((.(....).)))))))...... ( -32.10, z-score = -2.45, R) >consensus ____UUUUA___GGGCGGACAAUUUCAUCAAUGUCUUUGCCGCCAC__UGUAAAUGCUCGAGGGACAGAUACACAUGCCCACUUGGCCACAGCAGCGGCCAAGACAAUA .............(((((.(((...((....))...)))))))).................(((.((........))))).(((((((.(....).)))))))...... (-21.31 = -23.17 + 1.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:50 2011