| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,190,850 – 2,190,949 |
| Length | 99 |
| Max. P | 0.708682 |

| Location | 2,190,850 – 2,190,949 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 57.72 |
| Shannon entropy | 0.56585 |
| G+C content | 0.41878 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -5.79 |
| Energy contribution | -6.59 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2190850 99 + 22422827 AUCCGCAACCAAAAGGAGGAAGGAUGAACAUUGCAUGUGUGUACAUACAUAUGUACAUACGCAGUACGU-----------AGUACGGAUUAC---GUAUUACGUAUGGGCAGU ((((.(..((....))..)..))))....(((((..((((((((((....))))))))))...((((((-----------((((((.....)---)))))))))))..))))) ( -37.10, z-score = -4.84, R) >droYak2.chrX 15635102 98 - 21770863 AUCCACAACAAAAAGGAAGAAG-ACAAACAUUGUAUGUAUAUACAAACGUGCAUACAUACGUAGUACGU-----------AGUAGGGAUUAC---GUAGUACGUACGGACAGU .(((..................-........(((((((((........)))))))))((((((.(((((-----------(((....)))))---))).)))))).))).... ( -28.60, z-score = -4.26, R) >droPer1.super_26 892486 110 - 1349181 AAGCGUAUCGACCAAGAGGUAC-ACAGUUGUUG-GUGCAUGUUGUUGCAU-UGUCCACACGAAUUCCCUUCAGGCCAGAGAGCAGCGAUUGCUCGAUUAUACGUAUACAUAAU ..(((((((((((((.((.(..-..).)).)))-))(((.((((((((.(-(((....))))....((....)).......))))))))))).)))...)))))......... ( -24.90, z-score = 0.29, R) >consensus AUCCGCAACAAAAAGGAGGAAG_ACAAACAUUG_AUGUAUGUACAUACAU_UGUACAUACGAAGUACGU___________AGUAGGGAUUAC___GUAAUACGUAUGGACAGU .............................(((((((((((((....)))))))).((((((.(((((............................))))).)))))).))))) ( -5.79 = -6.59 + 0.80)


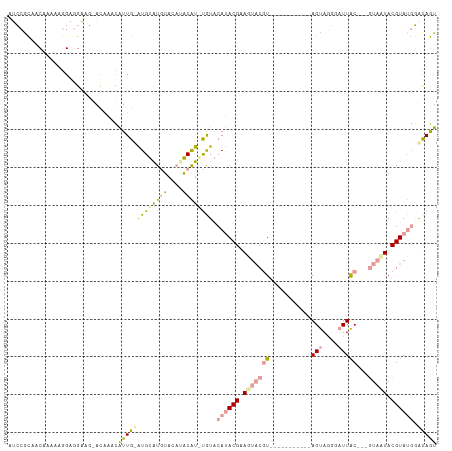
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:47 2011