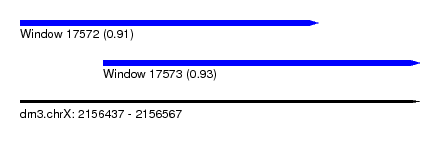
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,156,437 – 2,156,567 |
| Length | 130 |
| Max. P | 0.925849 |

| Location | 2,156,437 – 2,156,534 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.39605 |
| G+C content | 0.44880 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.82 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
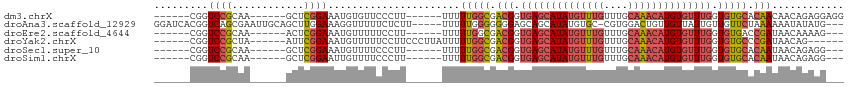
>dm3.chrX 2156437 97 + 22422827 ------CGGUCCGCAA------GCUCGGAAAUGUGUUCCCUU------UUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAACAACAGAGGAGG ------...((((...------...))))......((((((.------((.(((((.(((.((((((((((((((....)))))))))))))).))))).))).)).)).)))). ( -32.30, z-score = -1.72, R) >droAna3.scaffold_12929 2056366 106 + 3277472 GGAUCACGGUCAGCGAAUUGCAGCUUGGAAAGGUUUUUCUCUU-----UUUUUGGGGGGGAGCAGCAUAUGUGC-CGUGGACUGUUGUUAUUGUUGUUCUAAAAAAUAUAUG--- .(((.((((((.(((...(((.((((....))))((..((((.-----.....))))..)))))(((....)))-))).)))))).))).......................--- ( -26.10, z-score = -0.56, R) >droEre2.scaffold_4644 2121850 94 + 2521924 ------CGGUCCGCAA------ACUCGGAAAUGUUUUUCCUU------UUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGACCGAUAACAAAAG--- ------((((((((..------....(((((....)))))..------......)))(((.((((((((((((((....)))))))))))))).))))))))..........--- ( -34.14, z-score = -3.97, R) >droYak2.chrX 15598192 97 - 21770863 ------CGGUCCGCUA------AUUCGGAAAUGUUUUUCCUUCCCUUAUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCCCGAUAACAG------ ------(((..(((((------(...(((((....)))))...........))))))(((.((((((((((((((....)))))))))))))).)))..))).......------ ( -30.54, z-score = -2.91, R) >droSec1.super_10 1942271 94 + 3036183 ------CGGUCCGCAA------GCUCGGAAAUGUUUUCCCUU------UUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG--- ------...((((...------...)))).........((((------(..(((((.(((.((((((((((((((....)))))))))))))).))))).)))....)))))--- ( -30.30, z-score = -2.00, R) >droSim1.chrX 1515303 94 + 17042790 ------CGGUCCGCAA------GCUCGGAAUUGUUUUCCCUU------UUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG--- ------...((((...------...)))).........((((------(..(((((.(((.((((((((((((((....)))))))))))))).))))).)))....)))))--- ( -30.30, z-score = -2.05, R) >consensus ______CGGUCCGCAA______GCUCGGAAAUGUUUUCCCUU______UUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG___ .........((((............))))......................(((((.(((.((((((((((((((....)))))))))))))).))))).)))............ (-16.70 = -17.82 + 1.12)

| Location | 2,156,464 – 2,156,567 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.32858 |
| G+C content | 0.46063 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2156464 103 + 22422827 ------CUUUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAACAACAGAGGAGGCACUUGCCACGAUAACCUACUAACCGCGACGUC----- ------........((((.(((((((((((((((((....)))))))))))))(((((((.(..(......)..)))))..)))..............))))...))))----- ( -30.50, z-score = -1.14, R) >droEre2.scaffold_4644 2121877 104 + 2521924 ------CUUUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGACCGAUAACAAAAG---CACCUGCCACGUCUAUCGA-UAACUGACUAACCAAGUU ------.....(((((....((((((((((((((((....)))))))))))))(((((..............)---)))).)))..(((......-.....)))...))))).. ( -29.04, z-score = -1.83, R) >droYak2.chrX 15598219 107 - 21770863 CUUCCCUUAUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCCCGAUAAC---AG---CACCUGCCACGUCUAUCGA-UAACCGACUGACCAGCUC .............(((((.....(((((((((((((....)))))))))))))(((((..........---.)---))))))))).(((..(((.-....)))..)))...... ( -32.40, z-score = -2.52, R) >droSec1.super_10 1942298 98 + 3036183 ------CUUUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG---CACCUGCCACGAUAACCGACUAAUCC--ACGUC----- ------((.((.(((((.(((.((((((((((((((....)))))))))))))).))))).))).)).)).((---(....))).........(((......--..)))----- ( -29.10, z-score = -1.50, R) >droSim1.chrX 1515330 98 + 17042790 ------CUUUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG---CACCUGCCACGAUAACCGACUAAUCC--ACGUC----- ------((.((.(((((.(((.((((((((((((((....)))))))))))))).))))).))).)).)).((---(....))).........(((......--..)))----- ( -29.10, z-score = -1.50, R) >consensus ______CUUUUUUUGGCGACGGUGAGCAUAUGUUUGUUUGCAAACAUGUGUUUGGUGUGCACAAUAACAGAGG___CACCUGCCACGAUAACCGACUAACCG__ACGUC_____ .............(((((..((((((((((((((((....))))))))))))...(((........))).......)))))))))............................. (-24.34 = -24.38 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:43 2011