| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,154,129 – 2,154,232 |
| Length | 103 |
| Max. P | 0.699291 |

| Location | 2,154,129 – 2,154,232 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.55787 |
| G+C content | 0.42495 |
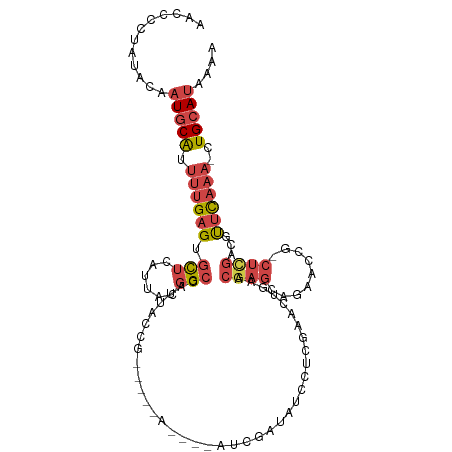
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -4.46 |
| Energy contribution | -5.55 |
| Covariance contribution | 1.09 |
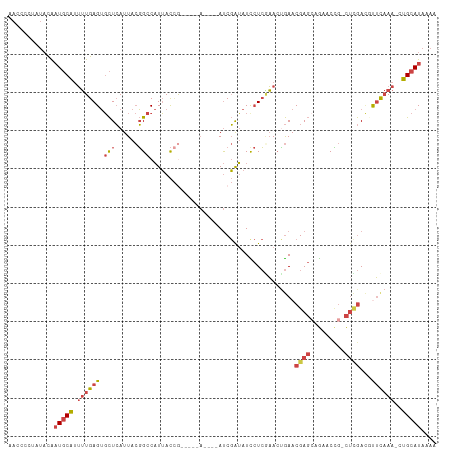
| Combinations/Pair | 1.29 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
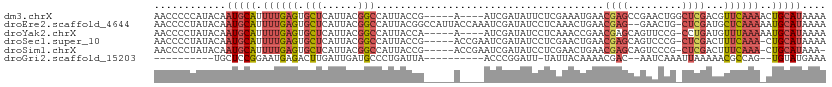
>dm3.chrX 2154129 103 + 22422827 AACCCCCAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACCG-----A----AUCGAUAUUCUCGAAAUGAACGAGCCGAACUGGCUCGACGUUCAAAACUGCAUAAAA ............((((((((((((((.(((((.(((......)))-----.----.((((.....))))))))).((((((.....)))))).))))))))).))))).... ( -29.70, z-score = -4.45, R) >droEre2.scaffold_4644 2119440 109 + 2521924 AACCCCUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACGGCCAUUACCAAAUCGAUAUCCUCAAACUGAACGAG--GAACUG-CUCGAUGCUCAAAAAUGCAUAAAA ............(((((((((((((.........((((.....))))........(((((..(((((.........)))--))....-.))))))))).))))))))).... ( -31.00, z-score = -5.11, R) >droYak2.chrX 15595879 102 - 21770863 AACCCCUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACCA-----A----AUCGAUAUCCUCAAACCGAACGAGCAGUUCCG-CCUGAUGUUUAAAAAUGCAUAAAA ............(((((((((.......(((((.(((........-----.----...((.....)).....((((.....)))).)-)))))))....))))))))).... ( -19.30, z-score = -2.16, R) >droSec1.super_10 1939983 105 + 3036183 AACCCCUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACCG-----ACCGAAUCGAUAUCCUCGAACUGAACGAGCAGUCCCG-CUCGACUUUCAAA-CUGCAUAAAA ............(((((.(((((((((((....(((......)))-----......((((.....)))).......)))))(((...-...))).))))))-.))))).... ( -25.00, z-score = -3.63, R) >droSim1.chrX 1513062 104 + 17042790 AACCCCUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACCG-----ACCGAAUCGAUAUCCUCGAACUGAACGAGCAGUCCCG-CUCGACUUUCAAA-CUGCAUAAA- ............(((((.(((((((((((....(((......)))-----......((((.....)))).......)))))(((...-...))).))))))-.)))))...- ( -25.00, z-score = -3.62, R) >droGri2.scaffold_15203 1546772 87 + 11997470 ----------UGCUCCGGAAUGAGACUUGAUUGAUGCCCUGAUUA----------ACCCGGAUU-UAUUACAAAACGAC--AAUCAAAUUAAAAACGCCAG--UGUAUGAAA ----------...((((((((.((..............)).))).----------..)))))((-(((.(((...((..--..............))....--)))))))). ( -7.93, z-score = 1.20, R) >consensus AACCCCUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACCG_____A____AUCGAUAUCCUCGAACUGAACGAGCAGAACCG_CUCGACGUUCAAA_CUGCAUAAAA ............(((((..(((((.(((......)))......................................((((.........))))...)))))...))))).... ( -4.46 = -5.55 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:41 2011