
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,143,591 – 2,143,711 |
| Length | 120 |
| Max. P | 0.589700 |

| Location | 2,143,591 – 2,143,711 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Shannon entropy | 0.40811 |
| G+C content | 0.49944 |
| Mean single sequence MFE | -33.21 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.39 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
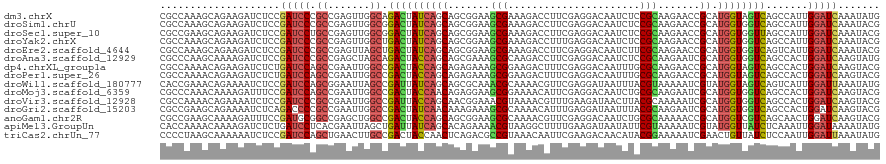
>dm3.chrX 2143591 120 - 22422827 CGCCAAAGCAGAAGAUCUCCGAUCCCGCCGAGUUGGCAGACUAUCAGCAGCGGAAGCGAAAGACCUUCGAGGACAAUCUCCGCAAGAACCGCAUGGUAGUCAGCCAUUGGAUCAAAUAUG .((....))...........(((((.(((.....))).((((((((...((((...(....)..(((((..(.....)..)).)))..)))).)))))))).......)))))....... ( -35.50, z-score = -1.56, R) >droSim1.chrU 15720461 120 - 15797150 CGCCAAAGCAGAAGAUCUCCGAUCCCGCCGAGUUGGCGGACUAUCAGCAGCGGAAGCGAAAGACCUUCGAGGACAAUCUCCGCAAGAACCGCAUGGUGGUCAGCCAUUGGAUCAAAUACG .((....))...........(((((((((.....))))((((((((...((((...(....)..(((((..(.....)..)).)))..)))).)))))))).......)))))....... ( -37.00, z-score = -0.89, R) >droSec1.super_10 1929519 120 - 3036183 CGCCGAAGCAGAAGAUCUCCGAUCCUGCCGAGUUGGCGGACUAUCAGCAGCGGAAGCGAAAGACCUUCGAGGACAAUCUCCGCAAGAACCGCAUGGUGGUUAGCCAUUGGAUCAAAUACG ..((((.((((..((((...)))))))).....((((.((((((((...((((...(....)..(((((..(.....)..)).)))..)))).)))))))).)))))))).......... ( -37.10, z-score = -0.76, R) >droYak2.chrX 15585173 120 + 21770863 CGCCAAAGCAGAAGAUCUCCGAUCCCGCCGAGUUGGCUGACUAUCAGCAGCGGAAGCGAAAGACCUUUGAGGACAAUCUCCGCAAGAACCGCAUGGUGGUCAGCCAUUGGAUCAAAUACG .((....))...........(((((........(((((((((((((...((((...(....)...((((.(((.....))).))))..)))).)))))))))))))..)))))....... ( -41.30, z-score = -2.54, R) >droEre2.scaffold_4644 2108675 120 - 2521924 CGCCAAAGCAGAAGAUCUCCGAUCCCGCCGAGUUAGCUGACUAUCAGCAGCGGAAGCGAAAGACCUUCGAGGACAAUCUUCGCAAGAACCGCAUGGUGGUCAGUCAUUGGAUCAAAUACG .((....))....(((((..(((.((((((.....((((.....)))).((((...(....)..((((((((.....))))).)))..)))).))))))...)))...)))))....... ( -37.90, z-score = -1.93, R) >droAna3.scaffold_12929 1242015 120 + 3277472 CGCCCAAGCAAAAGAUCUCCGAUCCCGCCGAGCUAGCAGACUACCAGCAGCGAAAGCGCAAGACCUUCGAGGACAAUCUCCGCAAGAAUCGCAUGGUGGUCAGCCACUGGAUCAAGUAUG .((....))...........(((((.(..(.(((....((((((((((.((....)).......(((((..(.....)..)).)))....)).)))))))))))).).)))))....... ( -33.60, z-score = -0.83, R) >dp4.chrXL_group1a 158754 120 - 9151740 CGCCAAAACAGAAGAUCUCUGAUCCAGCCGAAUUGGCCGACUACCAGCAGAGAAAGCGGAAGACUUUCGAGGACAAUUUGCGCAAGAACCGCAUGGUAGUCAGCCACUGGAUCAAGUACG ...................((((((((......((((.((((((((((...((((((....).)))))............(....)....)).)))))))).))))))))))))...... ( -42.40, z-score = -4.23, R) >droPer1.super_26 836950 120 + 1349181 CGCCAAAACAGAAGAUCUCUGAUCCAGCCGAAUUGGCCGACUACCAGCAGAGAAAGCGGAAGACUUUCGAGGACAAUUUGCGCAAGAACCGCAUGGUAGUCAGCCACUGGAUCAAGUACG ...................((((((((......((((.((((((((((...((((((....).)))))............(....)....)).)))))))).))))))))))))...... ( -42.40, z-score = -4.23, R) >droWil1.scaffold_180777 4167443 120 + 4753960 CACCGAAACAGAAAAUCUCCGAUCCAGCGGAAUUAGCCGAUUAUCAGCAGCGCAAACGCAAAACGUUCGAGGAUAAUUUACGUAAAAAUCGUAUGGUAGUCAGUCAUUGGAUUAAAUAUG ....................(((((((.((......))((((((((((.(((....)))...((((.............)))).......)).)))))))).....)))))))....... ( -22.12, z-score = -0.05, R) >droMoj3.scaffold_6359 4167195 120 - 4525533 CGCCCAAACAAAAGAUUUCCGAUCCAGCCGAAUUGGCCGACUACCAACAGAGGAAGCGGAAAACAUUCGAGGACAAUCUGCGCAAGAAUCGCAUGGUGGUCAGCCACUGGAUCAAGUACG ....................(((((((......((((.((((((((.((((.....((((.....)))).......)))).((.......)).)))))))).)))))))))))....... ( -36.40, z-score = -2.38, R) >droVir3.scaffold_12928 5642771 120 - 7717345 CGCCAAAACAGAAAAUCUCCGAUCCCGCCGAAUUGGCCGAUUACCAGCAACGGAAACGUAAAACGUUUGAAGAUAACUUACGCAAAAAUCGCAUGGUGGUCAGCCACUGGAUCAAGUACG ....................(((((.(......((((.((((((((((.(((....)))....(((..(.......)..)))........)).)))))))).))))).)))))....... ( -31.60, z-score = -2.01, R) >droGri2.scaffold_15203 1525632 120 - 11997470 CGCCGAAGCAGAAAAUCUCAGACCCCGCCGAAUUGGCCGACUAUCAACAAAGAAAGCGCAAAACAUUUGAGGAUAAUUUACGCAAGAAUCGCAUGGUGGUCAGCCACUGGAUCAAGUACG ..((...((......(((((((....(((.....)))((.((.((......)).)))).......)))))))........(....)....))..(((((....))))))).......... ( -23.50, z-score = 0.69, R) >anoGam1.chr2R 11800200 120 - 62725911 CGCCGAAGCAAAAGAUUUCCGAUGCGGCCGAGCUGGCCGACUACCAGCAGCGGAAGCGCAAAACGUUCGAGGACAAUCUGCGCAAAAACCGCAUGGUCGUCAGCAACUGGAUCAAGUACG .((....))....((..((((.(((((((.....))))(((.((((...((((..(((((....(((....)))....))))).....)))).)))).))).)))..))))))....... ( -39.10, z-score = -0.88, R) >apiMel3.GroupUn 11163918 120 - 399230636 CACCAAAACAAAAGAUCUCUGAUCCUCACGAAUUAGCUGAUUAUCAGCACAGAAAACGUAAGGCUUUUGAAGAUAAUAUUCGUAAAAAUCGUAUGGUUAUCUCAAAUUGGAUAAAAUAUG ..((((..((((((...((((.((.....))....((((.....)))).))))...(....).)))))).(((((((...((.......))....)))))))....)))).......... ( -23.30, z-score = -1.98, R) >triCas2.chrUn_77 78013 120 + 283703 CCCCUAAGCAAAAAAUCUCCGAUCCAGCUGAACUUGCCGACUACCAACUCAGACGCCGUAAACAAUUCGAAGACAACAUACGGAAAAAUCGAACUGUUAUCUCCAAUUGGAUUAAAUAUG ....................(((((((((((..(((........))).))))........((((.(((((......(....)......))))).))))........)))))))....... ( -15.00, z-score = -0.08, R) >consensus CGCCAAAGCAGAAGAUCUCCGAUCCCGCCGAAUUGGCCGACUAUCAGCAGCGGAAGCGAAAGACCUUCGAGGACAAUCUCCGCAAGAACCGCAUGGUGGUCAGCCAUUGGAUCAAAUACG ....................(((((.((.......)).((((((((((.......(((......................))).......)).)))))))).......)))))....... (-18.55 = -18.39 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:40 2011