
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,137,675 – 2,137,787 |
| Length | 112 |
| Max. P | 0.849769 |

| Location | 2,137,675 – 2,137,787 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Shannon entropy | 0.27790 |
| G+C content | 0.52281 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -27.71 |
| Energy contribution | -28.83 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
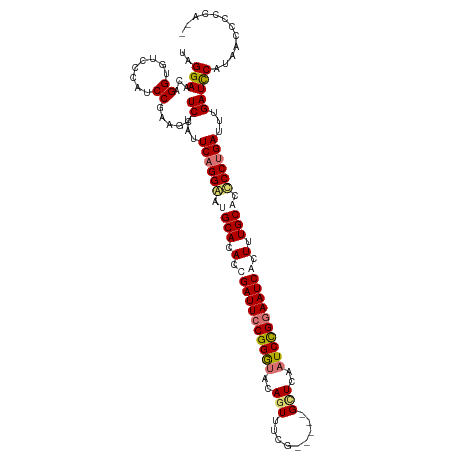
>dm3.chrX 2137675 112 + 22422827 UUGGAACAGGUGUCCCAUCCGAAGC-UCUAUUCAGGGAGGCACACCGAUUCCGGGUACAGUUUCG-----GCUUAAUCCGGAAUCACUUUGCACCCCUGAUUUGAUCCGUAACCCCCA-- ..(((...((........)).....-((...((((((..(((....((((((((((..(((....-----)))..))))))))))....)))..))))))...)))))..........-- ( -37.40, z-score = -1.99, R) >droEre2.scaffold_4644 2102718 113 + 2521924 UAGGAACAGGUGUCCCAUCCAGAGCAUCUCUUCAGGAAUGCACACCGAUUCCGGGUACAUUUUCG-----GUUCGAUCUGGAAUCGCUUUGCAGUCCUGAUUUGAUCCAUAACCCCCA-- ..(((.((((((..(......)..))))...((((((.((((...(((((((((((..((.....-----))...)))))))))))...)))).))))))..)).)))..........-- ( -37.00, z-score = -2.53, R) >droYak2.chrX 15579114 115 - 21770863 UUGGAACAGGUGUCCCAUCCGCAGCAUCUCUUCAGGAAUGCACAGCGAUUCCGGACACAGUCUCG-----GUUCGAUCUGAAAUCGCUUUGCAGCCCCGAUUUGAUUCACAAUCCCCCCA ..(((..((((((..(....)..))))))..((.((..((((.(((((((((((((...))).))-----)..........))))))).))))..)).))............)))..... ( -28.40, z-score = 0.08, R) >droSec1.super_10 1923665 117 + 3036183 UAGGAACAGGUGUCCCAUCCGAAGU-UCUAUUCAGGGGCGCACACCGAUUCCGGGUACAGUACCGUACCGGCUUAAUCCGGAAUCACUUUGCACCCCUGAUUUGAUCCGUAACCCCCA-- ..(((...((........)).....-((...(((((((.(((....((((((.(((((......)))))((......))))))))....))).)))))))...)))))..........-- ( -40.20, z-score = -2.44, R) >consensus UAGGAACAGGUGUCCCAUCCGAAGC_UCUAUUCAGGAAUGCACACCGAUUCCGGGUACAGUUUCG_____GCUCAAUCCGGAAUCACUUUGCACCCCUGAUUUGAUCCAUAACCCCCA__ ..(((...((........))......((...((((((.((((.(.(((((((((((...................))))))))))).).)))).))))))...)))))............ (-27.71 = -28.83 + 1.13)

| Location | 2,137,675 – 2,137,787 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Shannon entropy | 0.27790 |
| G+C content | 0.52281 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -35.33 |
| Energy contribution | -35.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2137675 112 - 22422827 --UGGGGGUUACGGAUCAAAUCAGGGGUGCAAAGUGAUUCCGGAUUAAGC-----CGAAACUGUACCCGGAAUCGGUGUGCCUCCCUGAAUAGA-GCUUCGGAUGGGACACCUGUUCCAA --..((((((....((....(((((((.(((.(.(((((((((....((.-----.....))....))))))))).).))).)))))))))..)-)))))...((((((....)))))). ( -41.30, z-score = -1.51, R) >droEre2.scaffold_4644 2102718 113 - 2521924 --UGGGGGUUAUGGAUCAAAUCAGGACUGCAAAGCGAUUCCAGAUCGAAC-----CGAAAAUGUACCCGGAAUCGGUGUGCAUUCCUGAAGAGAUGCUCUGGAUGGGACACCUGUUCCUA --((((..(..((..(((..((((((.((((.(.(((((((...(((...-----)))....(....)))))))).).)))).))))))((((...))))...)))..))...)..)))) ( -35.00, z-score = -0.71, R) >droYak2.chrX 15579114 115 + 21770863 UGGGGGGAUUGUGAAUCAAAUCGGGGCUGCAAAGCGAUUUCAGAUCGAAC-----CGAGACUGUGUCCGGAAUCGCUGUGCAUUCCUGAAGAGAUGCUGCGGAUGGGACACCUGUUCCAA ........(((((.(((...(((((..((((.(((((((((.....)).(-----((.(((...))))))))))))).))))..)))))...)))..))))).((((((....)))))). ( -37.80, z-score = -0.53, R) >droSec1.super_10 1923665 117 - 3036183 --UGGGGGUUACGGAUCAAAUCAGGGGUGCAAAGUGAUUCCGGAUUAAGCCGGUACGGUACUGUACCCGGAAUCGGUGUGCGCCCCUGAAUAGA-ACUUCGGAUGGGACACCUGUUCCUA --..((((((....((....(((((((((((.(.(((((((((......))((((((....)))))).))))))).).)))))))))))))..)-)))))....(((((....))))).. ( -50.60, z-score = -3.08, R) >consensus __UGGGGGUUACGGAUCAAAUCAGGGCUGCAAAGCGAUUCCAGAUCAAAC_____CGAAACUGUACCCGGAAUCGGUGUGCAUCCCUGAAGAGA_GCUUCGGAUGGGACACCUGUUCCAA ....(((((.....(((...(((((((((((.(.(((((((((.......................))))))))).).)))))))))))...))))))))....(((((....))))).. (-35.33 = -35.83 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:39 2011