| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,122,958 – 2,123,059 |
| Length | 101 |
| Max. P | 0.564829 |

| Location | 2,122,958 – 2,123,059 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Shannon entropy | 0.38115 |
| G+C content | 0.58125 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -22.40 |
| Energy contribution | -21.75 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
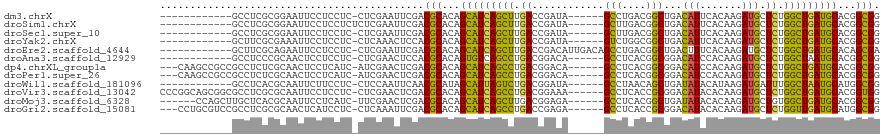
>dm3.chrX 2122958 101 + 22422827 ------------GCCUCGCGGAAUUCCUCCUC-CUCGAAUUCGACGCACAGCAUCAGCUUGACCGAUA------GCCUGACGGCUGACAUUCACAAGAUGCUCUGGCUGAUGCACGGCGG ------------(((..(((((((((......-...))))))..)))...(((((((((.((....((------(((....))))).((((.....)))).)).)))))))))..))).. ( -36.10, z-score = -2.51, R) >droSim1.chrX 1486059 102 + 17042790 ------------GCCUCGCGGAAUUCCUCCUCUCUCGAAUUCGACGCACAGCAUCAGCUUGACCGAUA------GCUUGACGGCUGACAUUCACAAGAUGCUCUGGCUGAUGCACGGCGG ------------(((..(((((((((..........))))))..)))...(((((((((.((....((------(((....))))).((((.....)))).)).)))))))))..))).. ( -33.50, z-score = -1.68, R) >droSec1.super_10 1908769 101 + 3036183 ------------GCCUCGCGGAAUUCCUCCUC-CUCGAAUUCGACGCACAGCAUCAGCUUGACCGAUA------GCUUGACGGCUGACAUUCACAAGAUGCUCUGGCUGAUGCACGGCGG ------------(((..(((((((((......-...))))))..)))...(((((((((.((....((------(((....))))).((((.....)))).)).)))))))))..))).. ( -33.60, z-score = -1.79, R) >droYak2.chrX 15564415 101 - 21770863 ------------GCUUCGCGAAAUUCCUCCUC-CUCAAACUCCACGCACAGCAUCAGCUUGACCGAUA------GUCUGGCGGCUGACAUUCACAAGAUGCUCUGGCUGAUGCACGGCGG ------------....................-...........(((...(((((((((.((....((------(((....))))).((((.....)))).)).)))))))))...))). ( -25.70, z-score = 0.22, R) >droEre2.scaffold_4644 2088089 107 + 2521924 ------------GCUUCGCAGAAUUCCUCCUC-CUCGAAUUCGACGCACAGCAUCAGCUUGACCGACAUUGACAGCCUGACGGCUGACUUUCACAAGAUGCUCUGGCUGAUGCACAGCGA ------------...((((.((((((......-...))))))........(((((((((.((.((.......(((((....))))).(((....))).)).)).)))))))))...)))) ( -34.90, z-score = -2.51, R) >droAna3.scaffold_12929 2021365 101 + 3277472 ------------GCCUCCCGCAACUCCUCCUC-CUCGAACUCCACGCACAGUGUCAGCCUGACGGACA------GCCUCACGGCGGACAUCCACAAGAUGCUCUGGCUAAUGCACGGCGG ------------.....((((...........-.............((.((((((.((((((.((...------.))))).)))((....))....)))))).))((....))...)))) ( -25.80, z-score = 0.49, R) >dp4.chrXL_group1a 136721 110 + 9151740 ---CAAGCCGCCGCCUCUCGCAACUCCUCAUC-AUCGAACUCGACGCACAGCAUCAGCCUGACGGACA------GCCUCACGGCGGACAUCCACAAGAUGCUCUGGCUGAUGCACGGCGG ---....((((((...................-.(((....)))......(((((((((.........------(((....)))(((((((.....)))).)))))))))))).)))))) ( -40.10, z-score = -2.66, R) >droPer1.super_26 814788 110 - 1349181 ---CAAGCCGCCGCCUCUCGCAACUCCUCAUC-AUCGAACUCGACGCACAGCAUCAGCCUGACGGACA------GCCUCACGGCGGACAUCCACAAGAUGCUCUGGCUGAUGCACGGCGG ---....((((((...................-.(((....)))......(((((((((.........------(((....)))(((((((.....)))).)))))))))))).)))))) ( -40.10, z-score = -2.66, R) >droWil1.scaffold_181096 8756827 101 - 12416693 ------------GCCUCACGCAAUUCUUCCUC-CUCCAAUUCAACGCAUAGCAUUAGUCUGACGGAUA------GCCUAACAGCUGAUAUACAUAAGAUGAUUUGGCUAAUGCAUGGCGG ------------((.....))...........-...........(((...(((((((((.......((------((......)))).....(((...)))....)))))))))...))). ( -19.90, z-score = 0.33, R) >droVir3.scaffold_13042 881704 113 + 5191987 CCCGGCAGCGGCGCCUCGCGCAAUUCCUCCUC-CUCGAACUCGACGCACAGCAUCAGCCUGACGGAAA------GCCUCACCGCGGACAUACACAAGAUGCUCUGGCUGAUGCACGGUGG .(((...(((((((...))))...........-.(((....))))))...((((((((((((.((...------.)))))....((((((.......))).)))))))))))).)))... ( -36.90, z-score = -0.35, R) >droMoj3.scaffold_6328 2872919 107 + 4453435 ------CCAGCUUGCUCACGCAAUUCCUCAUC-UUCGAACUCGACGCACAGCAUCAGCUUGACGGAGA------GCCUCACGGCUGAUAUACACAAGAUGCUGUGGCUGAUGCACGGCGG ------...(((((((((.((...........-.(((....)))..(((((((((..(((....)))(------(((....))))...........)))))))))))))).))).))).. ( -33.70, z-score = -1.13, R) >droGri2.scaffold_15081 1261715 110 + 4274704 ---CCUGCGUCCGCCUCGCGCAACUCAUCCUC-CUCAAAUUCGACGCACAGCAUCAGCCUGACCGAGA------GCCUCACCGCGGACAUACACAAGAUGCUCUGGUUGAUGCAUGGCGG ---.......(((((..((((...........-.........).)))...(((((((((......(((------((.((...(.(......).)..)).))))))))))))))..))))) ( -31.65, z-score = -0.39, R) >consensus ____________GCCUCGCGCAAUUCCUCCUC_CUCGAACUCGACGCACAGCAUCAGCCUGACCGAUA______GCCUCACGGCUGACAUUCACAAGAUGCUCUGGCUGAUGCACGGCGG ............................................(((...(((((((((.((............(((....)))...(((.......))).)).)))))))))...))). (-22.40 = -21.75 + -0.65)
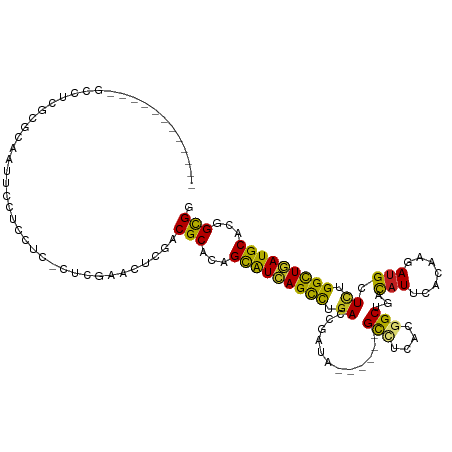

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:37 2011