| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,116,829 – 2,116,932 |
| Length | 103 |
| Max. P | 0.917672 |

| Location | 2,116,829 – 2,116,932 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Shannon entropy | 0.44310 |
| G+C content | 0.41715 |
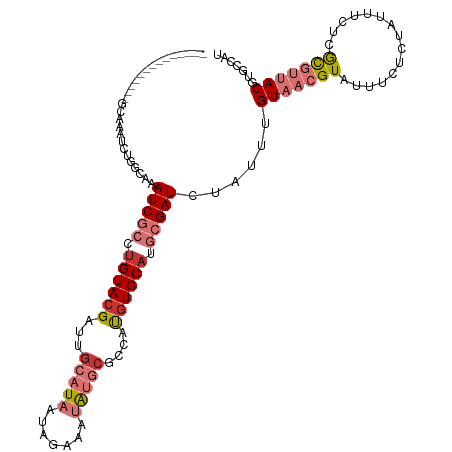
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -13.64 |
| Energy contribution | -15.26 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
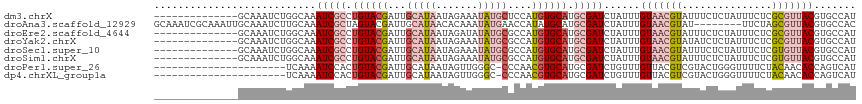
>dm3.chrX 2116829 103 + 22422827 --------------GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAAAUAUGCUCCAUGUGCAUGCGAUCUAUUUGUAACGUAUUUCUCUAUUUCUCGCGUUACGUGCCAU --------------.......(((((.(((((.((((((...(((((.......)))))....)))))).)))))......(((((((...............))))))).))))). ( -30.46, z-score = -3.55, R) >droAna3.scaffold_12929 2015179 109 + 3277472 GCAAAUCGCAAAUUGCAAAUCUUGCAAAUCGCUAGUACGAUUGCAUAACACAAAUAUGAACCAUAUGCAUGCGAUCUAUUUGUAACGUAU--------UUCUAGCGUUACGUGCCAC (((.((((((...(((......(((((.(((......))))))))........(((((...))))))))))))))......(((((((..--------.....))))))).)))... ( -26.70, z-score = -1.84, R) >droEre2.scaffold_4644 2081894 103 + 2521924 --------------GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAUAUAUGCGCCAUGUGCAUGCGAUCUAUUUGUAACGUAUUUCUCUAUUUCUCGCGUUACGUGCCAU --------------.......(((((.(((((.((((((..((((((.......))))))...)))))).)))))......(((((((...............))))))).))))). ( -30.86, z-score = -2.74, R) >droYak2.chrX 15558304 103 - 21770863 --------------GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAAAUAUGCGCCAUGUGCAUGCGAUCUAUUUGUAACGUAUAUCUCUAUUUCUCGCGUUACGUGCCAU --------------.......(((((.(((((.((((((..((((((.......))))))...)))))).)))))......(((((((...............))))))).))))). ( -30.86, z-score = -2.79, R) >droSec1.super_10 1902626 103 + 3036183 --------------GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAAAUAUGCGCCAUGUGCAUGCGAUCUAUUUGUAACGUAUUUCUCUAUUUCUCGUGUUACGUGCCAU --------------.......(((((.(((((.((((((..((((((.......))))))...)))))).)))))......((((((.................)))))).))))). ( -27.33, z-score = -1.96, R) >droSim1.chrX 1479885 103 + 17042790 --------------GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAAAUAUGCGCCAUGUGCAUGCGAUCUAUUUGUAACGUAUUUCUCUAUUUCUCGUGUUACGUGCCAU --------------.......(((((.(((((.((((((..((((((.......))))))...)))))).)))))......((((((.................)))))).))))). ( -27.33, z-score = -1.96, R) >droPer1.super_26 808778 94 - 1349181 ----------------------UCAAAAUCCACUGUACGAUUGCAUAAUAGUUGGGC-CCCAACGUGCAUGCGAUCUGUUUGUUACGUCGUACUGGGUUUUCUACAACACCAGUCAU ----------------------..((((((((..((((((((((((....(((((..-.)))))....)))))))).((.....))...))))))))))))................ ( -25.00, z-score = -1.45, R) >dp4.chrXL_group1a 130770 94 + 9151740 ----------------------UCAAAAUCCACUGUACGAUUGCAUAAUAGUUGGGC-CCCAACGUGCAUGCGAUCUGUUUGUUACGUCGUACUGGGUUUUCUACAACACCAGUCAU ----------------------..((((((((..((((((((((((....(((((..-.)))))....)))))))).((.....))...))))))))))))................ ( -25.00, z-score = -1.45, R) >consensus ______________GCAAAUCUGGCAAAUCGCCUGUACGAUUGCAUAAUAGAAAUAUGCGCCAUGUGCAUGCGAUCUAUUUGUAACGUAUUUCUCUAUUUCUCGCGUUACGUGCCAU ...........................(((((.((((((...(((((.......)))))....)))))).)))))......(((((((...............)))))))....... (-13.64 = -15.26 + 1.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:35 2011