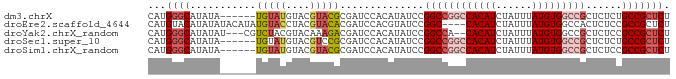
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,101,144 – 2,101,225 |
| Length | 81 |
| Max. P | 0.957675 |

| Location | 2,101,144 – 2,101,225 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 86.56 |
| Shannon entropy | 0.23041 |
| G+C content | 0.51987 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
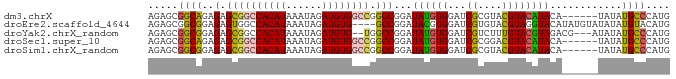
>dm3.chrX 2101144 81 + 22422827 AGAGCGGCAGAGAGCGGCCACAUAAAUAGAUGUGGCCGGCCGGAUAUGUGGAUCGCGUACGUACAUACA------UAUAUGCCCAUG ...(.((((.....(((((((((......))))))))).....(((((((...((....))....))))------))).)))))... ( -28.50, z-score = -2.01, R) >droEre2.scaffold_4644 2064139 83 + 2521924 AGAGCGGCGGAGAGUGGCCACAUAAAUAGAUGUG----GCCGGAUACGUGGAUCGUGUACGUAGGUACAUAUGUAUAUAUGUACAUG .((.(.(((.....(((((((((......)))))----))))....))).).))(((((((((.((((....)))).))))))))). ( -33.90, z-score = -4.78, R) >droYak2.chrX_random 199467 82 + 1802292 AGAGCGGCGGAGAGCGGCCACAUAAAUAGAUGUG--UGGCCGGAUAUGUGGAUCGUCUUUGUACGUAGACG---AUAUAUGCCCAUG ...(.(((......((((((((((......))))--)))))).....(((.(((((((........)))))---)).)))))))... ( -31.70, z-score = -3.39, R) >droSec1.super_10 1884538 81 + 3036183 AGAGCGGCAGAGAGCGGCCACAUAAAUAGAUGUGGCCGGCCGGAUAUGUGGAUCGCGGACGUACAUACA------UAUAUGCCCAUG ...(.((((.....(((((((((......))))))))).....(((((((...((....))....))))------))).)))))... ( -28.50, z-score = -2.06, R) >droSim1.chrX_random 1199170 81 + 5698898 AGAGCGGCGGAGAGCGGCCACAUAAAUAGAUGUGGCCGGCCGGAUAUGUGGAUCGCGUACGUACAUACA------UAUAUGCCCAUG ...(.(((((....(((((((((......))))))))).))..(((((((...((....))....))))------)))..))))... ( -29.10, z-score = -1.74, R) >consensus AGAGCGGCGGAGAGCGGCCACAUAAAUAGAUGUGGCCGGCCGGAUAUGUGGAUCGCGUACGUACAUACA______UAUAUGCCCAUG .(.(((((......(((((((((......))))))))))))...((((((...((....)))))))).............)).)... (-17.32 = -17.88 + 0.56)
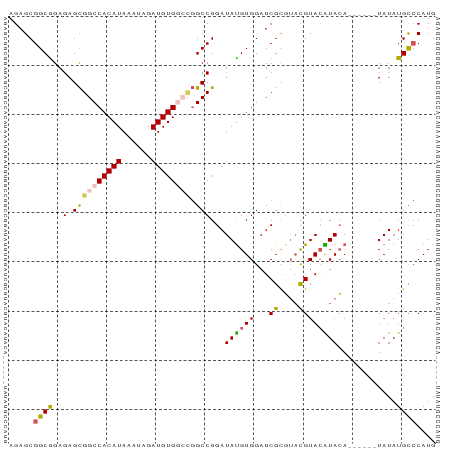
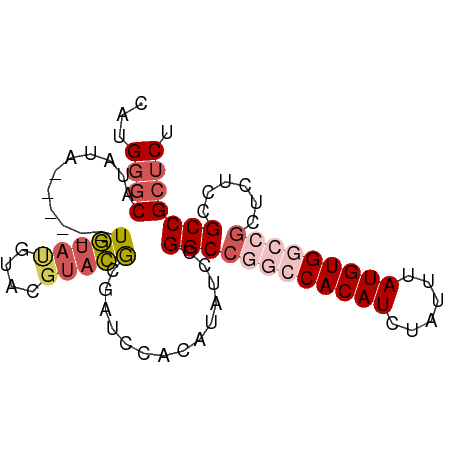
| Location | 2,101,144 – 2,101,225 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 86.56 |
| Shannon entropy | 0.23041 |
| G+C content | 0.51987 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.88 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
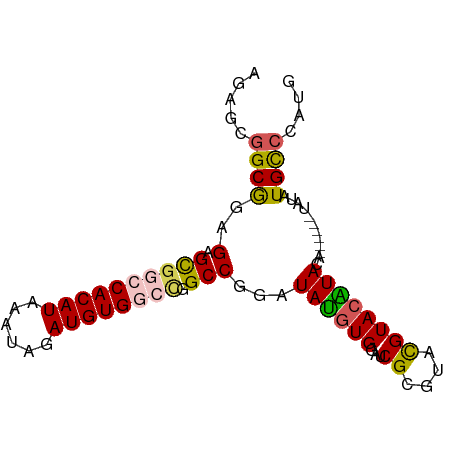
>dm3.chrX 2101144 81 - 22422827 CAUGGGCAUAUA------UGUAUGUACGUACGCGAUCCACAUAUCCGGCCGGCCACAUCUAUUUAUGUGGCCGCUCUCUGCCGCUCU ...((((..(((------(((..((.((....))))..))))))..((((((((((((......)))))))))......))))))). ( -28.50, z-score = -2.38, R) >droEre2.scaffold_4644 2064139 83 - 2521924 CAUGUACAUAUAUACAUAUGUACCUACGUACACGAUCCACGUAUCCGGC----CACAUCUAUUUAUGUGGCCACUCUCCGCCGCUCU ...((((((((....)))))))).(((((.........)))))...(((----(((((......))))))))............... ( -23.10, z-score = -3.84, R) >droYak2.chrX_random 199467 82 - 1802292 CAUGGGCAUAUAU---CGUCUACGUACAAAGACGAUCCACAUAUCCGGCCA--CACAUCUAUUUAUGUGGCCGCUCUCCGCCGCUCU ....(((....((---(((((........))))))).........((((((--((.(......).))))))))......)))..... ( -26.60, z-score = -3.95, R) >droSec1.super_10 1884538 81 - 3036183 CAUGGGCAUAUA------UGUAUGUACGUCCGCGAUCCACAUAUCCGGCCGGCCACAUCUAUUUAUGUGGCCGCUCUCUGCCGCUCU ...((((..(((------(((..((.((....))))..))))))..((((((((((((......)))))))))......))))))). ( -28.50, z-score = -2.39, R) >droSim1.chrX_random 1199170 81 - 5698898 CAUGGGCAUAUA------UGUAUGUACGUACGCGAUCCACAUAUCCGGCCGGCCACAUCUAUUUAUGUGGCCGCUCUCCGCCGCUCU ...((((..(((------(((..((.((....))))..))))))..((((((((((((......)))))))))......))))))). ( -28.50, z-score = -2.24, R) >consensus CAUGGGCAUAUA______UGUAUGUACGUACGCGAUCCACAUAUCCGGCCGGCCACAUCUAUUUAUGUGGCCGCUCUCCGCCGCUCU ...((((...........(((((....)))))..............((((((((((((......)))))))))......))))))). (-17.16 = -18.88 + 1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:33 2011