
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,089,124 – 2,089,229 |
| Length | 105 |
| Max. P | 0.992803 |

| Location | 2,089,124 – 2,089,229 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 72.25 |
| Shannon entropy | 0.46693 |
| G+C content | 0.51778 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
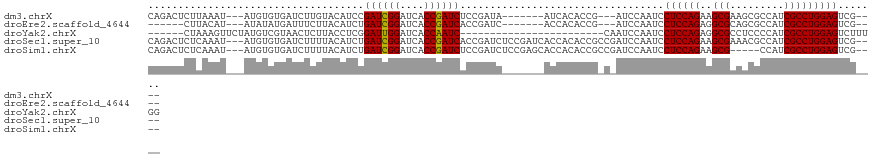
>dm3.chrX 2089124 105 + 22422827 CAGACUCUUAAAU---AUGUGUGAUCUUGUACAUCCGAUCGGAUCACCGAUCUCCGAUA-------AUCACACCG---AUCCAAUCCUCCAGAAGCGAAGCGCCAUCGCCUGGAGUCG---- ..(((........---..(((((((..(((......((((((....))))))....)))-------)))))))..---.........(((((..((((.......)))))))))))).---- ( -28.90, z-score = -2.09, R) >droEre2.scaffold_4644 2052327 99 + 2521924 ------CUUACAU---AUAUAUGAUUUCUUACAUCUGAUCGGAUCACCGAUCACCGAUC-------ACCACACCG---AUCCAAUCCUCCAGAGGCGCAGCGCCAUCGCCUGGAGUCG---- ------.......---......((((.........(((((((....)))))))..((((-------........)---))).))))((((((.((((...)))).....))))))...---- ( -25.00, z-score = -2.04, R) >droYak2.chrX 15536273 92 - 21770863 ------CUAAAGUUCUAUGUCGUAACUCUUACCUCGGAUUGGAUCACCAAUC------------------------CAAUCCAAUCCUCCAGAGGCGCCUCCCCAUCGCCUGGAGUCUUUGG ------((((((.......................(((((((((.....)))------------------------))))))....(((((..((((.........))))))))).)))))) ( -27.00, z-score = -3.31, R) >droSec1.super_10 1872411 115 + 3036183 CAGACUCUCAAAU---AUGUGUGAUCUUUUACAUCUGAUCGGAUCACCGAUCACCGAUCUCCGAUCACCACACCGCCGAUCCAAUCCUCCAGAAGCGAAACGCCAUCGCCUGGAGUCG---- ..(((........---.(((((((((.........(((((((....))))))).........))))).))))...............(((((..((((.......)))))))))))).---- ( -31.87, z-score = -2.65, R) >droSim1.chrX 1462996 110 + 17042790 CAGACUCUCAAAU---AUGUGUGAUCUUUUACAUCUGAUCGGAUCACCGAUCUCCGAUCUCCGAGCACCACACCGCCGAUCCAAUCCUCCAGAAGCG-----CCAUCGCCUGGAGUCG---- ..(((........---..(((((..(((....(((.((((((....))))))...)))....)))...)))))..............(((((..(((-----....))))))))))).---- ( -26.90, z-score = -1.03, R) >consensus CAGACUCUCAAAU___AUGUGUGAUCUUUUACAUCUGAUCGGAUCACCGAUCACCGAUC_______ACCACACCG_C_AUCCAAUCCUCCAGAAGCGAAGCGCCAUCGCCUGGAGUCG____ ....................................((((((....))))))..................................((((((..(((.........)))))))))....... (-18.82 = -18.50 + -0.32)

| Location | 2,089,124 – 2,089,229 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 72.25 |
| Shannon entropy | 0.46693 |
| G+C content | 0.51778 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -24.68 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
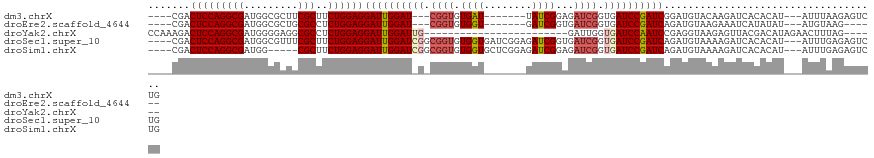
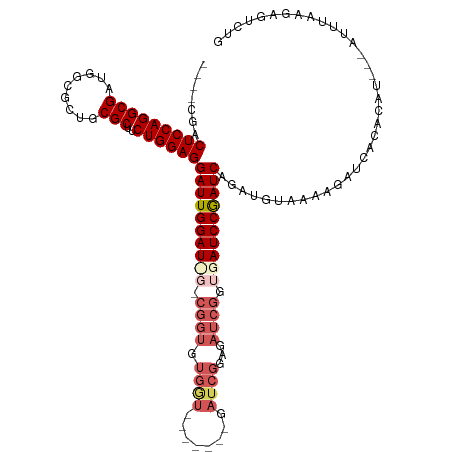
>dm3.chrX 2089124 105 - 22422827 ----CGACUCCAGGCGAUGGCGCUUCGCUUCUGGAGGAUUGGAU---CGGUGUGAU-------UAUCGGAGAUCGGUGAUCCGAUCGGAUGUACAAGAUCACACAU---AUUUAAGAGUCUG ----...((((((((((.......))))..))))))..((((((---..(((((((-------(..((..((((((....))))))...)).....))))))))..---))))))....... ( -35.90, z-score = -1.35, R) >droEre2.scaffold_4644 2052327 99 - 2521924 ----CGACUCCAGGCGAUGGCGCUGCGCCUCUGGAGGAUUGGAU---CGGUGUGGU-------GAUCGGUGAUCGGUGAUCCGAUCAGAUGUAAGAAAUCAUAUAU---AUGUAAG------ ----...(((((((....((((...)))))))))))..(((.((---..(((((((-------...((.(((((((....)))))))..))......)))))))..---)).))).------ ( -30.90, z-score = -0.49, R) >droYak2.chrX 15536273 92 + 21770863 CCAAAGACUCCAGGCGAUGGGGAGGCGCCUCUGGAGGAUUGGAUUG------------------------GAUUGGUGAUCCAAUCCGAGGUAAGAGUUACGACAUAGAACUUUAG------ ((.....(((((((((.........))))..)))))..((((((((------------------------((((...))))))))))))))..((((((.(......)))))))..------ ( -31.60, z-score = -2.37, R) >droSec1.super_10 1872411 115 - 3036183 ----CGACUCCAGGCGAUGGCGUUUCGCUUCUGGAGGAUUGGAUCGGCGGUGUGGUGAUCGGAGAUCGGUGAUCGGUGAUCCGAUCAGAUGUAAAAGAUCACACAU---AUUUGAGAGUCUG ----...((((((((((.......))))..))))))....(((((...((((((((((((..(.(((..(((((((....)))))))))).)....)))))).)))---)))...)).))). ( -42.20, z-score = -1.68, R) >droSim1.chrX 1462996 110 - 17042790 ----CGACUCCAGGCGAUGG-----CGCUUCUGGAGGAUUGGAUCGGCGGUGUGGUGCUCGGAGAUCGGAGAUCGGUGAUCCGAUCAGAUGUAAAAGAUCACACAU---AUUUGAGAGUCUG ----...(((((((((....-----)))..))))))((((...((((..(((((((.((...(.(((...((((((....)))))).))).)...)))))))))..---..)))).)))).. ( -36.50, z-score = -0.48, R) >consensus ____CGACUCCAGGCGAUGGCGCUGCGCUUCUGGAGGAUUGGAU_G_CGGUGUGGU_______GAUCGGAGAUCGGUGAUCCGAUCAGAUGUAAAAGAUCACACAU___AUUUAAGAGUCUG .......(((((((((.........)))..))))))((((((((((.((((.((((........))))...)))).)))))))))).................................... (-24.68 = -26.12 + 1.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:30 2011