| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,742,588 – 9,742,686 |
| Length | 98 |
| Max. P | 0.996085 |

| Location | 9,742,588 – 9,742,686 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 66.79 |
| Shannon entropy | 0.61424 |
| G+C content | 0.56492 |
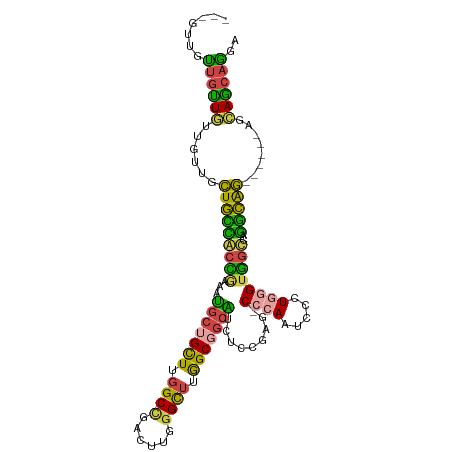
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -27.97 |
| Energy contribution | -26.42 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
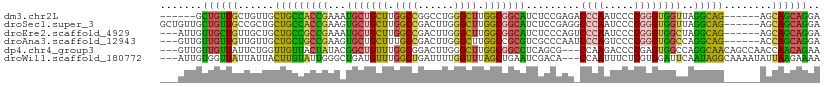
>dm3.chr2L 9742588 98 + 23011544 ------GCUGUUGCUGUUGCUGCCACCGAAAUGCUGCUUGGCCGGCCUGGGCUUGGCGGCAUCUCCGAGACCCAAUCCCUGGGUGGUUAGGCAG------AGCAGCAGGA ------....((((((((.((((((((((.((((((((.((((......)))).)))))))).))....(((((.....))))))))..)))))------)))))))).. ( -49.80, z-score = -2.44, R) >droSec1.super_3 5190082 104 + 7220098 GCUGUUGCUGUUGCCGCUGCUGCCACCGAAGUGCUGCUUGGCCGACUUGGGCUUGGCGGCAUCUCCGAGGCCCAAUCCCUGGGUGGUUAGGCAG------AGCAGCAGGA .((((((((.(((((......((((((((..(((((((.((((......)))).)))))))..))..(((.......))).))))))..)))))------)))))))).. ( -52.50, z-score = -1.85, R) >droEre2.scaffold_4929 10356550 101 + 26641161 ---AUUGUUGCUGUUGCUGCUGCCGCCGAAAUGCUGCUUGGCCGACUUGGGCUUGGCGGCAUCUCCCAGUCCCAAUCCCUGGGUGGCUAGGCAG------AGCAGCAGGA ---.......((((((((.(((((((((..((((((((.((((......)))).))))))))..(((((.........)))))))))..)))))------)))))))).. ( -52.00, z-score = -2.93, R) >droAna3.scaffold_12943 1171611 101 + 5039921 ---GUUGUUGUUGUUGUUGCUGCUGCCGAAGUGCUGCUUUGCCGACUUGGGCUUGGCCGCGUCGCCCAAUCCCAGUCCCUGGGUGGCCAGGCAG------ACCAGCAGGA ---.............(((((((((((.....((.(((..(((......)))..))).))((((((((...........))))))))..)))))------..)))))).. ( -40.60, z-score = -0.39, R) >dp4.chr4_group3 11266357 104 - 11692001 ---GUUGUUGUUAUUCUGGUUGUUACUAUACGGCUGUUUGGCGGACUUGGGCUUGGCGGCCUCAGCG---CCAAGACCCUGAGUGGCCAGGCAACAGCCAACCAACAGAA ---.((((((......((((....))))...(((((((((((.(.(((((((((((((.......))---))))).))).)))).))))...)))))))...)))))).. ( -42.60, z-score = -2.11, R) >droWil1.scaffold_180772 1094117 104 - 8906247 ---AUUGUGGUUAUUAUUACUUGUAUUGGGCUGAUGUUUGGCUGAUUUUGGUUUAGCUGAAUCGACA---CCAAUUUCUUGUGGAUUCAAUAGGCAAAAUAUUAAGAAAA ---................((((((((..(((.......((((((.......))))))(((((.(((---.........))).)))))....)))..)))).)))).... ( -21.00, z-score = -0.64, R) >consensus ___GUUGUUGUUGUUGUUGCUGCCACCGAAAUGCUGCUUGGCCGACUUGGGCUUGGCGGCAUCUCCGAG_CCCAAUCCCUGGGUGGCCAGGCAG______AGCAGCAGGA .......((((((......(((((((((...(((((((.((((......)))).))))))).........((((.....))))))))..)))))........)))))).. (-27.97 = -26.42 + -1.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:51 2011