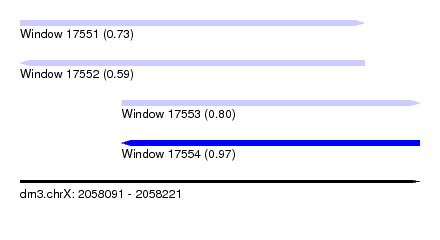
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,058,091 – 2,058,221 |
| Length | 130 |
| Max. P | 0.972679 |

| Location | 2,058,091 – 2,058,203 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.16781 |
| G+C content | 0.43978 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -23.96 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
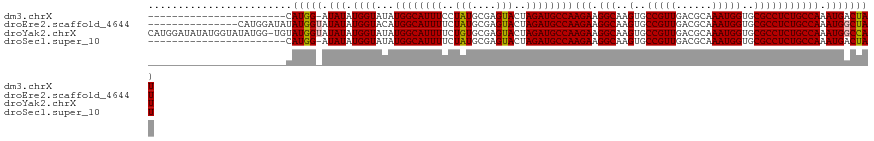
>dm3.chrX 2058091 112 + 22422827 -----ACACACAGACACUACAUCAUAACAAUAAAAAUAAUAGUCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGGAAAUGCC- -----....................................(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...))))- ( -21.80, z-score = -0.39, R) >droEre2.scaffold_4644 2021105 105 + 2521924 -------------GCACUACAUUAUAACAAUAAAAAUAAUAGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCCA -------------............................(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...)))). ( -27.10, z-score = -2.00, R) >droYak2.chrX 15504349 105 - 21770863 -------------GCACUACAUCAUAACAAUAAAAAUAAUGGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCACAGAAAAUGCCA -------------............................(((....)))...((((((.....))))))...((((..((((.....))))(((.((......)))))...)))). ( -25.50, z-score = -0.99, R) >droSec1.super_10 1841791 117 + 3036183 AGACUCUACGCAGACACUACAUCAUAACAAUAAAAAUAAUAGUCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCC- ....((((.((((((..........................(((....)))(((((((((.....)))))).........((((.....))))))).)).)).)).)))).......- ( -24.00, z-score = -0.53, R) >consensus _____________ACACUACAUCAUAACAAUAAAAAUAAUAGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCC_ .........................................(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...)))). (-23.96 = -23.78 + -0.19)

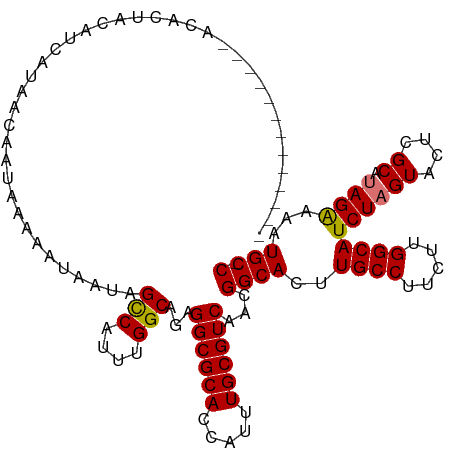
| Location | 2,058,091 – 2,058,203 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.16781 |
| G+C content | 0.43978 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2058091 112 - 22422827 -GGCAUUUCCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAUUAUUUUUAUUGUUAUGAUGUAGUGUCUGUGUGU----- -(((((((..(((....)))..))))))).(((.(((..(..(((((......)))))..)))))))((((...(((.(((((..((((....)))).))))))))..)).))----- ( -30.40, z-score = -0.90, R) >droEre2.scaffold_4644 2021105 105 - 2521924 UGGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGGCUAUUAUUUUUAUUGUUAUAAUGUAGUGC------------- ((((((((..(((....)))..))))))))(((.(((..(..(((((......)))))..)))))))(((....)))((((((..((((....)))).)))))).------------- ( -31.30, z-score = -1.40, R) >droYak2.chrX 15504349 105 + 21770863 UGGCAUUUUCUGUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGGCCAUUAUUUUUAUUGUUAUGAUGUAGUGC------------- ((((((((...(((....))).))))))))(((.(((..(..(((((......)))))..)))))))(((....)))((((((..((((....)))).)))))).------------- ( -33.50, z-score = -1.68, R) >droSec1.super_10 1841791 117 - 3036183 -GGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAUUAUUUUUAUUGUUAUGAUGUAGUGUCUGCGUAGAGUCU -(((....((((((((..(((((.((((..(((.(((..(..(((((......)))))..)))))))))...(((((.............))))).)).)))))..))))))))))). ( -35.22, z-score = -1.72, R) >consensus _GGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAUUAUUUUUAUUGUUAUGAUGUAGUGC_____________ .(((((((..(((....)))..)))))))....((((..(..(((((......)))))..)))))((((...(((((...((....))..)))))...))))................ (-27.94 = -27.50 + -0.44)

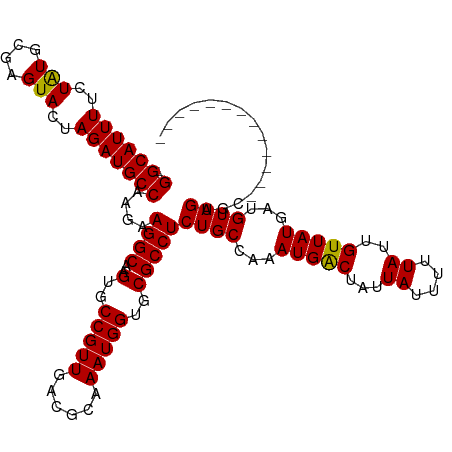
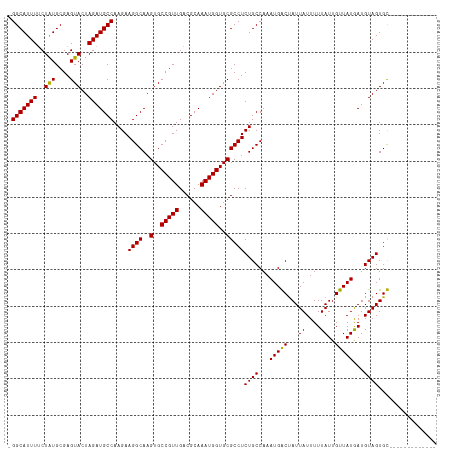
| Location | 2,058,124 – 2,058,221 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.22500 |
| G+C content | 0.45509 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2058124 97 + 22422827 AUAGUCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGGAAAUGCCAUAUACCAUAUAU-CCAUG----------------------- ...(..((.(((((..((((((.....))))))...(((....)))((((((((...........)).)))))).))))).))..)......-.....----------------------- ( -23.90, z-score = -0.67, R) >droEre2.scaffold_4644 2021130 106 + 2521924 AUAGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCCAUGUACCAUAUAUACCAUAUAUCCAUG--------------- ...(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...))))...........................--------------- ( -27.10, z-score = -1.22, R) >droYak2.chrX 15504374 120 - 21770863 AUGGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCACAGAAAAUGCCAUAUACCAUAUAUACCAUACA-CCAUAUACCAUAUAUCCAUG ((((..((.(((....((((((.....))))))...((((..((((.....))))(((.((......)))))...)))).....................-.......))).))..)))). ( -27.00, z-score = -0.81, R) >droSec1.super_10 1841829 97 + 3036183 AUAGUCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCCAUAUACCAUAUAU-CCAUG----------------------- ...(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...)))).............-.....----------------------- ( -24.40, z-score = -1.21, R) >consensus AUAGCCAUUUGGCAGAGGCGCACCAUUUGCGUCAACGGCACUUGCCUUCUUGGCAUCUAGUACUCGCAUAGAAAAUGCCAUAUACCAUAUAU_CCAUA_______________________ ...(((....)))...((((((.....))))))...((((..((((.....))))((((((....)).))))...)))).......................................... (-24.81 = -24.62 + -0.19)
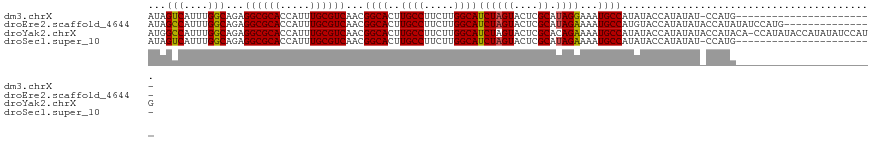
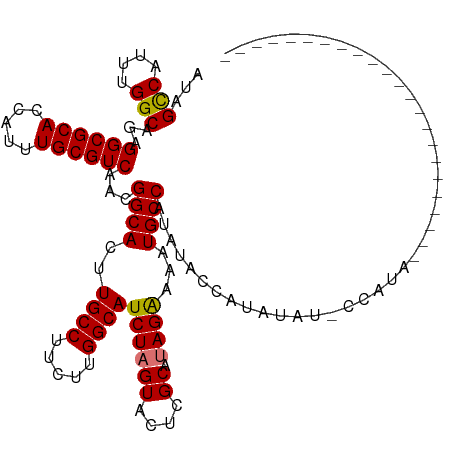
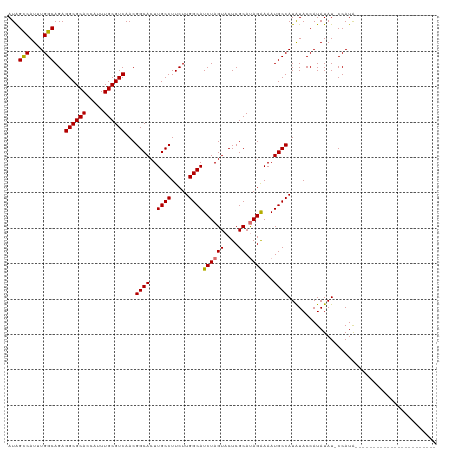
| Location | 2,058,124 – 2,058,221 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.22500 |
| G+C content | 0.45509 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.33 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
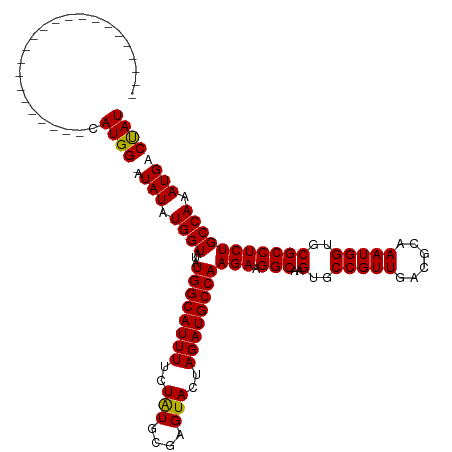
>dm3.chrX 2058124 97 - 22422827 -----------------------CAUGG-AUAUAUGGUAUAUGGCAUUUCCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAU -----------------------.((((-.(((.((((...((((((((..(((....)))..))))))))(((.(((..(..(((((......)))))..))))))))))).))).)))) ( -32.30, z-score = -2.23, R) >droEre2.scaffold_4644 2021130 106 - 2521924 ---------------CAUGGAUAUAUGGUAUAUAUGGUACAUGGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGGCUAU ---------------..........((((((...((((((.(.((((.....)))).))))))).))))))(((.(((..(..(((((......)))))..)))))))(((....)))... ( -37.80, z-score = -2.52, R) >droYak2.chrX 15504374 120 + 21770863 CAUGGAUAUAUGGUAUAUGG-UGUAUGGUAUAUAUGGUAUAUGGCAUUUUCUGUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGGCCAU .........(((((...(((-(...((((((...((((((.(.((((.....)))).))))))).))))))(((.(((..(..(((((......)))))..)))))))))))....))))) ( -41.10, z-score = -1.56, R) >droSec1.super_10 1841829 97 - 3036183 -----------------------CAUGG-AUAUAUGGUAUAUGGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAU -----------------------.((((-.(((.((((...((((((((..(((....)))..))))))))(((.(((..(..(((((......)))))..))))))))))).))).)))) ( -32.90, z-score = -2.54, R) >consensus _______________________CAUGG_AUAUAUGGUAUAUGGCAUUUUCUAUGCGAGUACUAGAUGCCAAGAAGGCAAGUGCCGUUGACGCAAAUGGUGCGCCUCUGCCAAAUGACUAU ........................(((((.(((.((((...((((((((..(((....)))..))))))))(((.(((..(..(((((......)))))..))))))))))).)))))))) (-31.20 = -31.33 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:26 2011