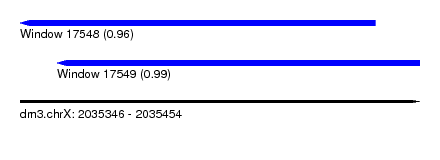
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,035,346 – 2,035,454 |
| Length | 108 |
| Max. P | 0.990939 |

| Location | 2,035,346 – 2,035,442 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.80 |
| Shannon entropy | 0.36225 |
| G+C content | 0.50011 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.66 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
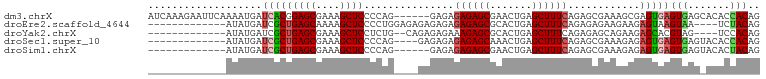
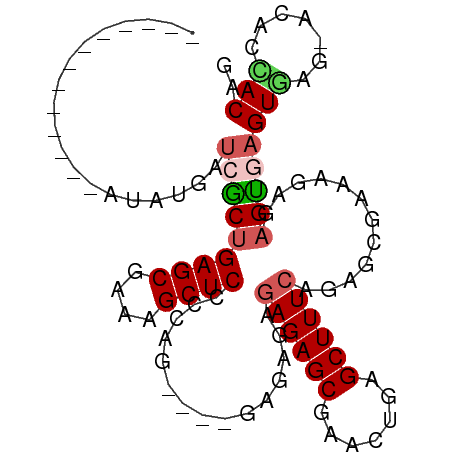
>dm3.chrX 2035346 96 - 22422827 AUCAAAGAAUUCAAAAUGAUCACGGAGCGAAAGCUCCCCAG------GAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGCGAGUGAGUGAGCACACCACAG .((................((..(((((....)))))....------....((((((.......)))))).))((....))))(((.(((....)))))).. ( -31.00, z-score = -3.77, R) >droEre2.scaffold_4644 1998561 85 - 2521924 -------------AUAUGAUCGCUGAGCAAAAGCUCCCCUGGAGAGAGAGAGAGAGCGCACUGAGCUUUCAGAGAGAAGAAGAGUAAGUAA----UCUACAG -------------....(((..((..((.....((((...)))).......((((((.......)))))).............)).))..)----))..... ( -17.00, z-score = 0.53, R) >droYak2.chrX 15481868 83 + 21770863 -------------AUAUGAUCGCUGAGCGAAAGCUCCUCUG--CAGAGAGAAAGAGCGCACUGAGCUUUCAGAGAGCAGAAGAGCACGUAG----UCCACAG -------------.((((.((((...))))..((((.((((--(.....(((((..(.....)..))))).....))))).)))).)))).----....... ( -24.10, z-score = -0.41, R) >droSec1.super_10 1818903 85 - 3036183 -------------AUAUGAUCGCUGAGCGAAAGCUCCCCAG----GAGAGAGAGAGCAAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAGUACACCACAG -------------......(((((((((....)))).....----......((((((.......))))))..)))))......(((.(((....)))))).. ( -28.00, z-score = -4.12, R) >droSim1.chrX 1417405 83 - 17042790 -------------AUAUGAUCGCUGAGCGAAAGCUCCCCAG------GAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAGUACACUACAG -------------......(((((((((....)))).....------....((((((.......))))))..)))))......((.((((....)))))).. ( -26.70, z-score = -3.22, R) >consensus _____________AUAUGAUCGCUGAGCGAAAGCUCCCCAG____GAGAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAG_ACACCACAG ...................(((((((((....))))...............((((((.......))))))............)))))(((.......))).. (-17.06 = -17.66 + 0.60)

| Location | 2,035,356 – 2,035,454 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.14 |
| Shannon entropy | 0.36347 |
| G+C content | 0.53273 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
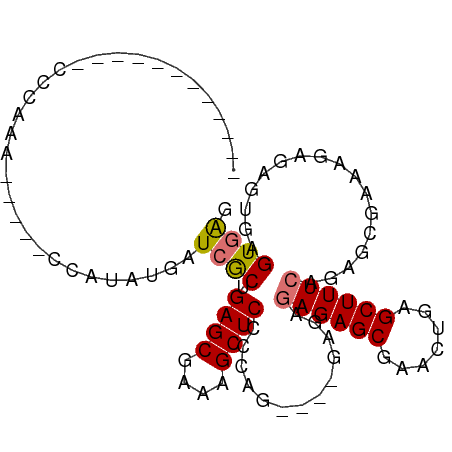
>dm3.chrX 2035356 98 - 22422827 CCCCCCCCCUCCAUCAAAGAAUUCAAAAUGAUCACGGAGCGAAAGCUCCCCAG----GAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGCGAGUGAGUGAG ........((((((((............))))...(((((....)))))...)----))).((((((.......))))))...((....))........... ( -30.40, z-score = -3.66, R) >droYak2.chrX 15481874 85 + 21770863 -----------CCCCAG------CCAUAUGAUCGCUGAGCGAAAGCUCCUCUGCAGAGAGAAAGAGCGCACUGAGCUUUCAGAGAGCAGAAGAGCACGUAGU -----------......------...((((.((((...))))..((((.(((((.....(((((..(.....)..))))).....))))).)))).)))).. ( -24.10, z-score = -0.78, R) >droSec1.super_10 1818913 83 - 3036183 ------------CCCAAA-----CCAUAUGAUCGCUGAGCGAAAGCUCCCCAG--GAGAGAGAGAGCAAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAG ------------.....(-----((((....(((((((((....)))).....--......((((((.......))))))..)))))......))).))... ( -22.70, z-score = -2.81, R) >droSim1.chrX 1417415 81 - 17042790 ------------CCCAAA-----CCAUAUGAUCGCUGAGCGAAAGCUCCCCAG----GAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAG ------------.....(-----((((....(((((((((....)))).....----....((((((.......))))))..)))))......))).))... ( -23.40, z-score = -2.98, R) >consensus ____________CCCAAA_____CCAUAUGAUCGCUGAGCGAAAGCUCCCCAG____GAGAGAGAGCGAACUGAGCUUUCAGAGCGAAAGAGAGUGAGUGAG ...............................((((.((((....)))).............((((((.......)))))).................)))). (-17.75 = -17.88 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:23 2011