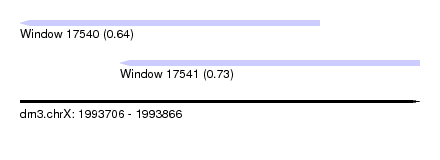
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,993,706 – 1,993,866 |
| Length | 160 |
| Max. P | 0.730026 |

| Location | 1,993,706 – 1,993,826 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Shannon entropy | 0.26335 |
| G+C content | 0.38917 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1993706 120 - 22422827 AGCCAAAAAUAUAUACCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUGUAACACUUUUUUGUAAGCAAAUCAAACCUCCAGCUCCGCA .((((((((..........((.(((((((....))))))).))......(((.((((((.((.((....)))))))))).)))....))))))..(((..............)))..)). ( -26.74, z-score = -2.23, R) >droSim1.chrX 1402428 96 - 17042790 AGCCAAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUGUAACACUUUUUUGUAA------------------------ ...((((((..........((.(((((((....))))))).))......(((.((((((.((.((....)))))))))).)))....))))))...------------------------ ( -24.90, z-score = -3.25, R) >droSec1.super_10 1778119 120 - 3036183 AGCCGAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUGUAACACUUUUUUGUAAGCAAAUCGGAUCUCCAGCUCCGCA .((.((.........((((((.(((((((....))))))).))......(((.((((((.((.((....)))))))))).)))......((((....)))))))).........)).)). ( -30.57, z-score = -2.17, R) >droYak2.chrX 15439691 120 + 21770863 AGCCAAAAAUAUCGCCCGAGAUUUACGAUCAGUGUCGUAAUUUAUAUUAUUAGAUGGAAUACUACGUGAGUGCGUCCAUGUAACAGUUUUUUCUAAGCAAAUCAGAUCUCCAGCUUCGCG .............((..((((((((((((....))))))..........(((.(((((....(((....)))..))))).)))..((((.....))))......))))))..))...... ( -24.70, z-score = -1.07, R) >droEre2.scaffold_4644 1966573 120 - 2521924 AGCCGAAAAUAUCUCCCGAGAUUUACGAUCAGUGCCGUAAUUUAUAUUAUUAGAUGGAACUCUACGUGAGUGCUGCCAUGUAACACUUUUUUGUAAGCAAAUCAGAUCUCCAGCUCCGCA .((.((....((((....))))....((((((((..(((((....)))))((.((((.((((.....))))....)))).)).))))..((((....))))...))))......)).)). ( -22.00, z-score = -0.00, R) >consensus AGCCAAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUGUAACACUUUUUUGUAAGCAAAUCAGAUCUCCAGCUCCGCA ...((((((..........((.(((((((....))))))).))......(((.((((((.((.((....)))))))))).)))....))))))........................... (-18.56 = -19.28 + 0.72)

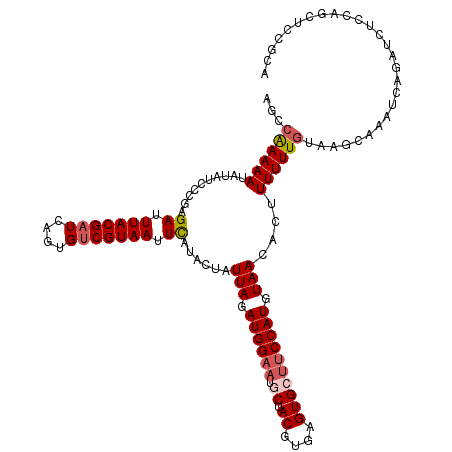
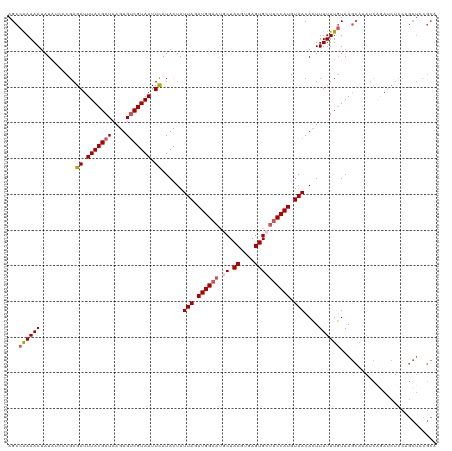
| Location | 1,993,746 – 1,993,866 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Shannon entropy | 0.11537 |
| G+C content | 0.41167 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1993746 120 - 22422827 CGAGAUUUAUGAUACUGCUCGAUUUUUGCCAUGCUCCACCAGCCAAAAAUAUAUACCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUG ...((.(((((((((((.((((((((((....(((.....))))))))))...............))).))))))))))).))..........((((((.((.((....)))))))))). ( -32.56, z-score = -3.33, R) >droSim1.chrX 1402444 120 - 17042790 CGAGAUUUAUGAUACUGCUCCAUUUUUGCCAUGCUCCACCAGCCAAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUG ...((.(((((((((((.((.(((((((....(((.....))))))))))..(((....)))....)).))))))))))).))..........((((((.((.((....)))))))))). ( -31.70, z-score = -3.35, R) >droSec1.super_10 1778159 120 - 3036183 CGAGAUUUAUGAUACUGCUCCAUUUUUGCCAUGCUCCACCAGCCGAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUG ...((.(((((((((((.((.(((((((....(((.....))))))))))..(((....)))....)).))))))))))).))..........((((((.((.((....)))))))))). ( -31.40, z-score = -3.02, R) >droYak2.chrX 15439731 120 + 21770863 CGAGAUUUAUGAUACUGCUACAGUUUUACCAUGCUCCGCCAGCCAAAAAUAUCGCCCGAGAUUUACGAUCAGUGUCGUAAUUUAUAUUAUUAGAUGGAAUACUACGUGAGUGCGUCCAUG ...((.(((((((((((...............(((.....)))...((((.((....)).)))).....))))))))))).))..........(((((....(((....)))..))))). ( -23.00, z-score = -0.18, R) >droEre2.scaffold_4644 1966613 120 - 2521924 CGAGAUUUAUGAUACUGCUCCAUUUUUCCCAUGCUCCACCAGCCGAAAAUAUCUCCCGAGAUUUACGAUCAGUGCCGUAAUUUAUAUUAUUAGAUGGAACUCUACGUGAGUGCUGCCAUG ...((.(((((.(((((.((...(((((....(((.....))).))))).((((....))))....)).))))).))))).))..........((((.((((.....))))....)))). ( -21.20, z-score = -0.38, R) >consensus CGAGAUUUAUGAUACUGCUCCAUUUUUGCCAUGCUCCACCAGCCAAAAAUAUAUCCCGAGAUUUACGAUCAGUGUCGUAAUUCAUACUAUUAGAUGGAAUGCUACGUGAGUGCUUCCAUG ...((.(((((((((((....(((((((....(((.....))))))))))......((.......))..))))))))))).))..........((((((.((.((....)))))))))). (-22.78 = -23.90 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:16 2011