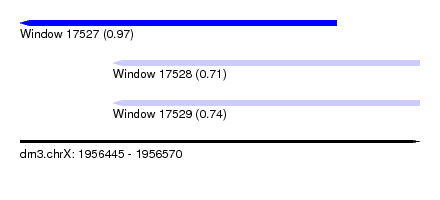
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,956,445 – 1,956,570 |
| Length | 125 |
| Max. P | 0.969520 |

| Location | 1,956,445 – 1,956,544 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.75 |
| Shannon entropy | 0.61061 |
| G+C content | 0.56482 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.63 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969520 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
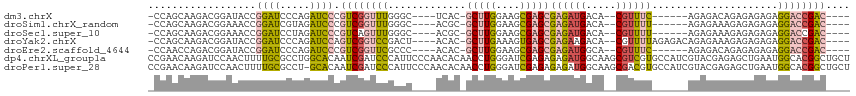
>dm3.chrX 1956445 99 - 22422827 -CCAGCAAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUUGGGC----UCAC-GCUUGGAAGCGAGCGAGAUGACA--CGUUUC------AGAGACAGAGAGAGAGGACCGAC---- -((........)).....(((((...))))).((((((((...(----((..-(((((....)))))((((((...--))))))------.........)))...))))))))---- ( -31.70, z-score = -1.58, R) >droSim1.chrX_random 1160173 99 - 5698898 -CCAGCAAGACGGAAACCGGAUCGUAGAUCCCGUCGGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA--CGUUUU------AGAGAAAGAGAGAGAGGACCGAC---- -..........(....).(((((...))))).((((((((...(----....-(((((....)))))((((((...--))))))------...........)...))))))))---- ( -25.20, z-score = -0.02, R) >droSec1.super_10 1741297 99 - 3036183 -CCAGCAAGACGGAAACCGGAUCCUAGAUCCCGUCAGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA--CGUUUU------AGAGAAAGAGAGAGAGGACCGAC---- -..........(....)(((.((((...((.(((((((....))----.(.(-(((((....)))))).).)))).--(.....------.).....).))...)))))))..---- ( -24.50, z-score = -0.25, R) >droYak2.chrX 15401301 105 + 21770863 -CCAGCAAGACGGAUACCGGAUCCCAGAUCCAGUCGGUCCGACU----ACAC-GCUUGAAAGUGAGCGAGAAGACA--CGUUUUAGAGACAGAGAAAGAGAGAGAGGACCGAC---- -((........)).....(((((...))))).((((((((..((----.(.(-((((......))))).).))...--.(((.....)))...............))))))))---- ( -28.70, z-score = -2.95, R) >droEre2.scaffold_4644 1929003 99 - 2521924 -CCAACCAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUCGCCC----ACAC-GCUUGGAAGCGAGCGAGAUGGCA--CGUUUC------AGAGACAGAGAGAGAGGACCGAC---- -....((....)).....(((((...))))).(((((((((((.----...(-(((((....))))))....))).--.(((..------...))).........))))))))---- ( -33.10, z-score = -2.41, R) >dp4.chrXL_group1a 2635551 117 - 9151740 CCGAACAAGAUCCAACUUUUGCGCCUGGCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAGCGUCGUGCCAUCGUACGAGAGCUGAAUGGCACGGCUGCU ...........(((.(((((((.....))....(((((((((..............)))))))))))))).)))..(((((((((((((....(....)....)))))))))).))) ( -40.64, z-score = -2.21, R) >droPer1.super_28 1072938 116 - 1111753 CCGAACAAGAUCCAACUUUUGCGCCU-GCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAGCGACGUGCCAUCGUACGAGAGCUGAAUGGCACGGCUGCU .....(((((......))))).(((.-......(((((((((..............))))))))).......))).((((.((((((((....(....)....))))))))..)))) ( -37.98, z-score = -2.34, R) >consensus _CCAGCAAGACGGAAACCGGAUCCCAGAUCCCGUCGGUUCGAGC____ACAC_GCUUGGAAGCGAGCGAGAUGACA__CGUUUU______AGAGAAAGAGAGAGAGGACCGAC____ ..................((((.....)))).((((((((.............(((((....)))))((((((.....)))))).....................)))))))).... (-15.64 = -15.63 + -0.01)
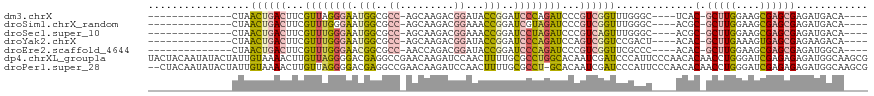
| Location | 1,956,474 – 1,956,570 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.24 |
| Shannon entropy | 0.57261 |
| G+C content | 0.54397 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
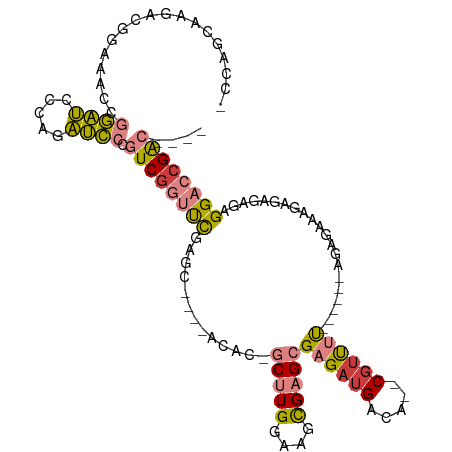
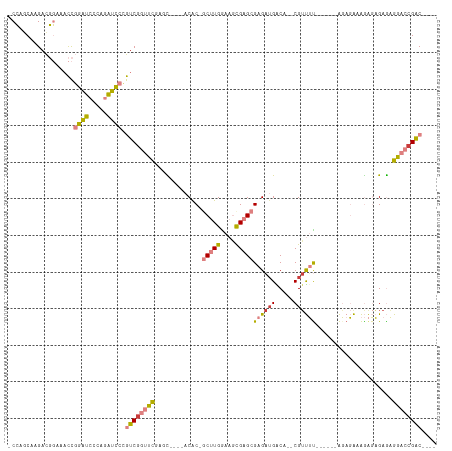
>dm3.chrX 1956474 96 - 22422827 --------------CUAACUGACUUCGUUAGGGAAUGGCGCC-AGCAAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUUGGGC----UCAC-GCUUGGAAGCGAGCGAGAUGACA---- --------------(((((((((...(((.((((.(((..((-........))...)))..)))).)))...))))).))))..----((.(-(((((....)))))).)).....---- ( -30.00, z-score = -0.10, R) >droSim1.chrX_random 1160202 96 - 5698898 --------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAAACCGGAUCGUAGAUCCCGUCGGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA---- --------------((((((((((((((((.((.......))-....)))))))....(((((...))))).))))).)))).(----((.(-(((((....)))))).).))...---- ( -29.70, z-score = 0.13, R) >droSec1.super_10 1741326 96 - 3036183 --------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAAACCGGAUCCUAGAUCCCGUCAGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA---- --------------(((((((((...((((((((.(((..((-........))...)))..))))))))...))))).)))).(----((.(-(((((....)))))).).))...---- ( -31.90, z-score = -0.95, R) >droYak2.chrX 15401336 96 + 21770863 --------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAUACCGGAUCCCAGAUCCAGUCGGUCCGACU----ACAC-GCUUGAAAGUGAGCGAGAAGACA---- --------------...(((((((..((((((((.(((..((-........))...)))..))))))))..)))))))....((----.(.(-((((......))))).).))...---- ( -30.80, z-score = -1.56, R) >droEre2.scaffold_4644 1929032 96 - 2521924 --------------CUAACUGACUUCGUUUGGGAACGGCGCC-AACCAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUCGCCC----ACAC-GCUUGGAAGCGAGCGAGAUGGCA---- --------------..(((((((...((((((((.(((..((-........))...)))..))))))))...))))))).(((.----...(-(((((....))))))....))).---- ( -38.90, z-score = -2.66, R) >dp4.chrXL_group1a 2635588 120 - 9151740 UACUACAAUAUACUAUUGUAAAACUUGUUAGGGGACGAGGCCGAACAAGAUCCAACUUUUGCGCCUGGCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAGCG ...(((((((...)))))))...(((((((.(((((((.((((..(((((......)))))....))))....))).))))..(((((........)))))((....))..))))))).. ( -32.90, z-score = -2.35, R) >droPer1.super_28 1072975 117 - 1111753 --CUACAAUAUACUAUUGUAAAACUUGUUAGGGGACGAGGCCGAACAAGAUCCAACUUUUGCGCCU-GCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAGCG --.(((((((...)))))))...((((((.((........)).))))))...........(((((.-......(((((((((..............))))))))).......)))..)). ( -30.98, z-score = -2.07, R) >consensus ______________CUAACUGACUUCGUUUGGGAAUGGCGCC_AGCAAGACGGAAACCGGAUCCCAGAUCCCGUCGGUUCGAGC____ACAC_GCUUGGAAGCGAGCGAGAUGACA____ .................((((((...((((((((.(((..................)))..))))))))...))))))...............(((((....)))))............. (-14.47 = -14.06 + -0.42)
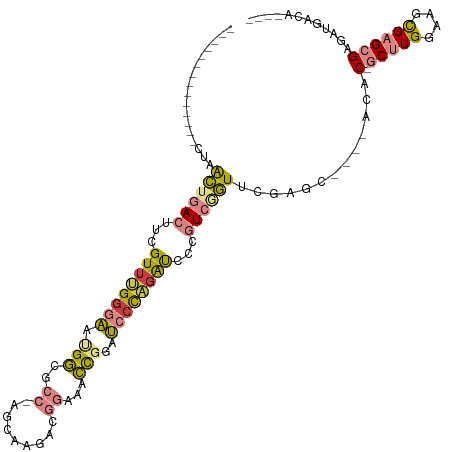
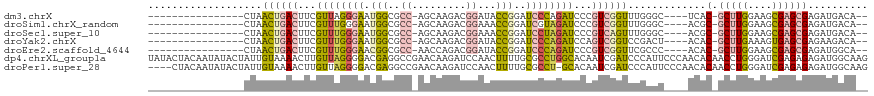
| Location | 1,956,474 – 1,956,570 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.44 |
| Shannon entropy | 0.56809 |
| G+C content | 0.54030 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1956474 96 - 22422827 ----------------CUAACUGACUUCGUUAGGGAAUGGCGCC-AGCAAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUUGGGC----UCAC-GCUUGGAAGCGAGCGAGAUGACA-- ----------------(((((((((...(((.((((.(((..((-........))...)))..)))).)))...))))).))))..----((.(-(((((....)))))).)).....-- ( -30.00, z-score = -0.10, R) >droSim1.chrX_random 1160202 96 - 5698898 ----------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAAACCGGAUCGUAGAUCCCGUCGGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA-- ----------------((((((((((((((((.((.......))-....)))))))....(((((...))))).))))).)))).(----((.(-(((((....)))))).).))...-- ( -29.70, z-score = 0.13, R) >droSec1.super_10 1741326 96 - 3036183 ----------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAAACCGGAUCCUAGAUCCCGUCAGUUUGGGC----ACGC-GCUUGGAAGCGAGCGAGAUGACA-- ----------------(((((((((...((((((((.(((..((-........))...)))..))))))))...))))).)))).(----((.(-(((((....)))))).).))...-- ( -31.90, z-score = -0.95, R) >droYak2.chrX 15401336 96 + 21770863 ----------------CUAACUGACUUCGUUUGGGAAUGGCGCC-AGCAAGACGGAUACCGGAUCCCAGAUCCAGUCGGUCCGACU----ACAC-GCUUGAAAGUGAGCGAGAAGACA-- ----------------...(((((((..((((((((.(((..((-........))...)))..))))))))..)))))))....((----.(.(-((((......))))).).))...-- ( -30.80, z-score = -1.56, R) >droEre2.scaffold_4644 1929032 96 - 2521924 ----------------CUAACUGACUUCGUUUGGGAACGGCGCC-AACCAGACGGAUACCGGAUCCCAGAUCCCGUCGGUUCGCCC----ACAC-GCUUGGAAGCGAGCGAGAUGGCA-- ----------------..(((((((...((((((((.(((..((-........))...)))..))))))))...))))))).(((.----...(-(((((....))))))....))).-- ( -38.90, z-score = -2.66, R) >dp4.chrXL_group1a 2635590 120 - 9151740 UAUACUACAAUAUACUAUUGUAAAACUUGUUAGGGGACGAGGCCGAACAAGAUCCAACUUUUGCGCCUGGCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAG .....(((((((...)))))))...(((((((.(((((((.((((..(((((......)))))....))))....))).))))..(((((........)))))((....))..))))))) ( -32.50, z-score = -2.45, R) >droPer1.super_28 1072977 115 - 1111753 ----CUACAAUAUACUAUUGUAAAACUUGUUAGGGGACGAGGCCGAACAAGAUCCAACUUUUGCGCCU-GCACAAUCGAUCCCAUUCCCAACACAACCUGGGAUCGAGAGAGAUGGCAAG ----.(((((((...)))))))...((((((.((........)).))))))..(((.(((((((....-))....(((((((((..............)))))))))))))).))).... ( -30.94, z-score = -2.38, R) >consensus ________________CUAACUGACUUCGUUUGGGAAUGGCGCC_AGCAAGACGGAAACCGGAUCCCAGAUCCCGUCGGUUCGAGC____ACAC_GCUUGGAAGCGAGCGAGAUGACA__ ...................((((((...((((((((.(((..................)))..))))))))...))))))...............(((((....)))))........... (-14.47 = -14.06 + -0.42)
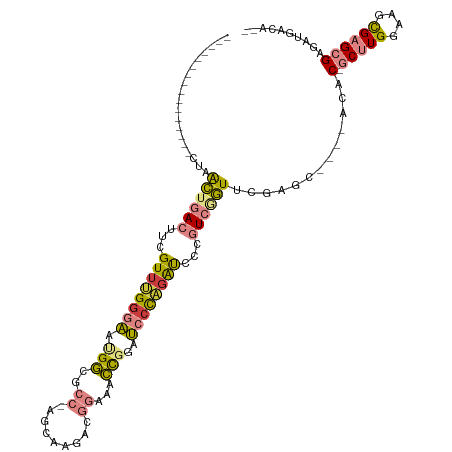
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:06 2011