
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,955,989 – 1,956,093 |
| Length | 104 |
| Max. P | 0.603990 |

| Location | 1,955,989 – 1,956,093 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Shannon entropy | 0.39525 |
| G+C content | 0.60542 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -16.33 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1955989 104 + 22422827 ------------UCCACCCACUCGCCCCUCA-AUUUCCGACCCUCA-CCAGCACACACAUGAGGGUUGCCAAGUGAGCGCC--AAGUGCCGAGCAUCGAGCAUGCGCCAACUUUUUCGCG ------------..........(((......-.....(((((((((-............)))))))))..((((..((((.--..((((((.....)).))))))))..))))....))) ( -29.60, z-score = -2.57, R) >droAna3.scaffold_12929 1954361 111 + 3277472 GGCAGCAAGAGGGUUGCCUAGUCACUCUUGGUGUGUGUGGCACUCAGUCGGGGCA-----AAGGGUUGUCGAGUGCGCCCCCAAACUGGAUGCCACUGAGCAUGCGCCAACAUGCG---- .(((.(((((((..(....)..).)))))).)))..((((((..((((.(((((.-----................)))))...))))..))))))...(((((......))))).---- ( -44.23, z-score = -0.43, R) >droEre2.scaffold_4644 1928509 101 + 2521924 ------------UACCCCCACCCGCCC-UCG-AUUUCCGACCCUCA-CCAGCACACA--UGAGGGUUGCCAAGUGAGCGCC--AAGUGCCGAGUACCGAGCAUGCGCCAACUUUUUCGCG ------------..........(((..-...-.....(((((((((-..........--)))))))))..((((..((((.--..((((((.....)).))))))))..))))....))) ( -29.40, z-score = -2.90, R) >droYak2.chrX 15400838 98 - 21770863 ------------UCCACCCACUCGCCCCUCG-AUUUCCGACCCUCA-CCAGCAC------GAGGGUUGCCAAGUGAGCGCC--AAGUGCCGAGCACCGAGCAUGCGCCAACUUUUUCGCG ------------..........(((......-.....((((((((.-.......------))))))))..((((..((((.--..((((((.....)).))))))))..))))....))) ( -29.40, z-score = -2.16, R) >droSec1.super_10 1740831 102 + 3036183 ------------UCCACCCACUCGCCCCUCA-AUUUCCGACCCUAA-CCAGCACACA--UGAGGGUUGCCAAGUAAGCGCC--AAGUGCCGAGCACCGAGCAUGCGCCAACUUUUUCGCG ------------..........(((......-.....(((((((..-..........--..)))))))..((((..((((.--..((((((.....)).))))))))..))))....))) ( -23.84, z-score = -1.26, R) >droSim1.chrX_random 1159717 102 + 5698898 ------------UCCACCCACUCGCCCCUCA-AUUUCCGACCCUAA-CCAGCACACA--UGAGGGUUGCCAAGUGAGCGCC--AAGUGCCGAGCACCGAGCAUGCGCCAACUUUUUCGCG ------------..........(((......-.....(((((((..-..........--..)))))))..((((..((((.--..((((((.....)).))))))))..))))....))) ( -24.84, z-score = -1.13, R) >consensus ____________UCCACCCACUCGCCCCUCA_AUUUCCGACCCUCA_CCAGCACACA__UGAGGGUUGCCAAGUGAGCGCC__AAGUGCCGAGCACCGAGCAUGCGCCAACUUUUUCGCG ......................(((............(((((((.................)))))))..((((..((((.....((((((.....)).))))))))..))))....))) (-16.33 = -17.58 + 1.25)

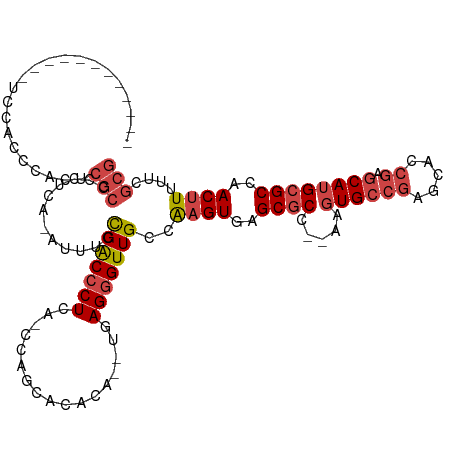

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:03 2011