| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,954,239 – 1,954,350 |
| Length | 111 |
| Max. P | 0.975551 |

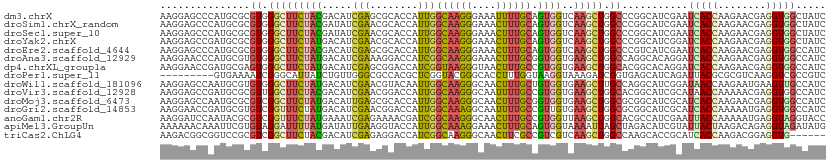
| Location | 1,954,239 – 1,954,350 |
|---|---|
| Length | 111 |
| Sequences | 15 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.92 |
| Shannon entropy | 0.54456 |
| G+C content | 0.54102 |
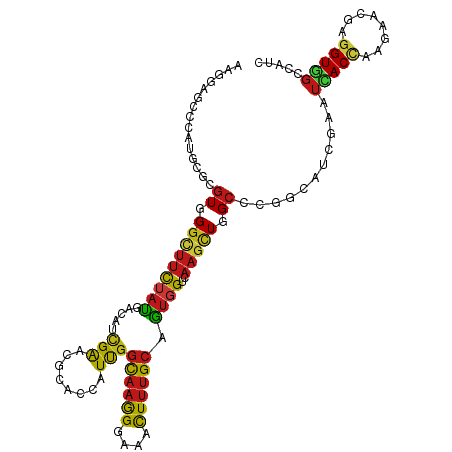
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1954239 111 - 22422827 AAGGAGCCCAUGCGCGUGGGCUUCUACGACAUCGAGCGCACCAUUGGCAAGGGAAAUUUUGCAGUGGUCAAGCUGGCCCGGCAUCGAAUCACCAAGAACGAGGUGGCUAUC .((((((((((....)))))))))).(((..(((.(((((((((((.(((((....))))))))))))...))..)).)))..)))..(((((........)))))..... ( -40.60, z-score = -1.35, R) >droSim1.chrX_random 1157962 111 - 5698898 AAGGAGCCCAUGCGCGUGGGCUUCUACGAUAUCGAACGCACCAUUGGCAAGGGAAACUUUGCAGUGGUCAAGCUGGCCCGGCAUCGAAUCACCAAGAACGAGGUGGCUAUC .((((((((((....))))))))))..((((((((..(.(((((((.(((((....)))))))))))))..((((...)))).)))).(((((........))))).)))) ( -46.90, z-score = -3.51, R) >droSec1.super_10 1739094 111 - 3036183 AAGGAGCCCAUGCGCGUGGGCUUCUACGAUAUCGAACGCACCAUUGGCAAGGGAAACUUUGCAGUGGUCAAGCUGGCCCGGCAUCGAAUCACCAAGAACGAGGUGGCUAUC .((((((((((....))))))))))..((((((((..(.(((((((.(((((....)))))))))))))..((((...)))).)))).(((((........))))).)))) ( -46.90, z-score = -3.51, R) >droYak2.chrX 15399016 111 + 21770863 AAGGAGCCGAUGCGCGUGGGCUUCUAUGACAUCGAACGCACCAUUGGCAAGGGAAACUUUGCAGUGGUCAAGCUGGCCCGGCAUCGGAUCACCAAGAACGAGGUGGCUAUC ......(((((((...((((((.((.((....))...(.(((((((.(((((....))))))))))))).))..))))))))))))).(((((........)))))..... ( -42.90, z-score = -2.17, R) >droEre2.scaffold_4644 1926689 111 - 2521924 AAGGAGCCCAUGCGCGUGGGCUUCUACGACAUCGAGCGCACCAUUGGCAAGGGAAACUUUGCAGUGGUCAAGCUGGCCCGUCAUCGAAUCACCAAGAACGAGGUGGCCAUC .((((((((((....)))))))))).((....))(((..(((((((.(((((....))))))))))))...)))(((((.((.((..........))..)).).))))... ( -47.80, z-score = -3.37, R) >droAna3.scaffold_12929 1952432 111 - 3277472 AAGGAACCCAUGCGUGUGGGCUUCUAUGACAUCGAAAGGACCAUCGGCAAGGGAAACUUUGCGGUGGUGAAGCUGGCCAGGCACAGGAUCACCAAGAACGAGGUGGCCAUC ..((..((..(((.(((.((((((.......((....))(((((((.(((((....)))))))))))))))))).).)).)))..)).(((((........)))))))... ( -45.90, z-score = -3.31, R) >dp4.chrXL_group1a 2632960 111 - 9151740 AAGGAACCGAUGCGAGUGGGCUUCUAUGACAUCGAGCGGACCAUCGGUAAGGGUAACUUUGCCGUGGUGAAGCUGGCACGGCACAGGAUCACCAAGAACGAGGUGGCCAUC ..((..((..((((.((.((((((..((....)).....((((.((((((((....)))))))))))))))))).)).).)))..)).(((((........)))))))... ( -40.10, z-score = -1.75, R) >droPer1.super_11 1365806 102 - 2846995 ---------GUGAAAAUCGGGCAUUAUCUGUUGGGCGCCACGCUCGGUACGGGCACCUUUGGUAAGGUAAAGAUCGGUGAGCAUCAGAUUACGCGCGUCAAGGUCGCCGUC ---------((((....((.((...(((((.((..((((..(((((...)))))(((((....))))).......))))..)).)))))...)).))......)))).... ( -33.70, z-score = -0.71, R) >droWil1.scaffold_181096 5477984 111 + 12416693 AAGGAGCCAAUGCGUGUGGGCUUCUAUGACAUCGAACGUACAAUUGGCAAGGGCAACUUUGCUGUGGUGAAGCUUGCCAGGCAUCGGAUAACCAAGAAUGAAGUUGCCAUC ..((..((.((((.((..((((((.((.(((.....((......))((((((....))))))))).))))))))..).).)))).))....)).................. ( -34.70, z-score = -0.90, R) >droVir3.scaffold_12928 7191329 111 - 7717345 AAGGAGCCGAUGCGCGUUGGCUUCUACGACAUCGAACGGACCAUUGGCAAGGGCAAUUUUGCCGUGGUGAAGCUGGCACGGCAUCGCAUAACCAAAAACGAGGUGGCCAUC ...(.(((((((((.((..(((((..((....)).....(((((.(((((((....)))))))))))))))))..)).).))))).....(((........)))))))... ( -42.10, z-score = -2.07, R) >droMoj3.scaffold_6473 14061921 111 + 16943266 AAGGAGCCAAUGCGCGUCGGCUUCUAUGACAUUGAGCGCACCAUUGGCAAGGGCAACUUUGCGGUGGUGAAGCUGGCGCGGCAUCGCAUCACCAAGAACGAGGUGGCCAUC ..((.((..(((((((((((((((...(........)..(((((((.(((((....))))))))))))))))))))))).)))).)).(((((........)))))))... ( -46.80, z-score = -2.33, R) >droGri2.scaffold_14853 1431330 111 - 10151454 AAGGAACCGAUGCGUGUCGGUUUCUAUGACAUCGAACGGACCAUUGGCAAAGGCAAUUUUGCCGUUGUGAAGCUGGCGCGGCAUCGCAUCACCAAAAAUGAGGUGGCCAUC ..((...(((((((((((((((((.(..((.((.....)).....(((((((....)))))))))..)))))))))))).))))))..(((((........)))))))... ( -43.00, z-score = -2.61, R) >anoGam1.chr2R 30992726 111 - 62725911 AAGGAUCCAAUACGCGUCGGUUUCUAUGAAAUCGAGAAAACGAUCGGCAAGGGCAACUUUGCCGUGGUUAAGCUGGCACGCCAUCGAAUUACCAAAAAUGAGGUAGGUACC ..((..((.....((.((((((((...))))))))...(((.(.((((((((....))))))))).)))..))(((....)))......((((........))))))..)) ( -33.80, z-score = -1.92, R) >apiMel3.GroupUn 23827977 111 - 399230636 AAAAAACAAAUUCGUGUAGGAUUUUAUGAUAUUGAAGGUACCAUUGGCAAAGGAAACUUUGCAGUGGUAAAAUUAGCUAGACAUCGUAUUACUAAGACAGAGGUAGAUAUG .............((.(((..(((((......))))).((((((((.(((((....))))))))))))).......))).))..(((((((((........))).)))))) ( -26.90, z-score = -3.25, R) >triCas2.ChLG4 4711961 105 - 13894384 AAGACGGCGGUCCGCGUCGGCUUCUACGACAUCGAGAGGACCAUCGGCAAGGGCAACUUCGCCGUCGUCAAGCUGGCCAAGCACCGCAUCACCAAGACGGAGGUG------ ......(((((.(..((((((((..(((((.((....))......(((.(((....))).)))))))).))))))))...).)))))..((((........))))------ ( -41.40, z-score = -1.73, R) >consensus AAGGAGCCCAUGCGCGUGGGCUUCUAUGACAUCGAACGCACCAUUGGCAAGGGAAACUUUGCAGUGGUCAAGCUGGCCCGGCAUCGAAUCACCAAGAACGAGGUGGCCAUC ...............((.(((((((((.....(((........)))((((((....)))))).))))..))))).))...........(((((........)))))..... (-17.99 = -17.50 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:02 2011