
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,931,455 – 1,931,553 |
| Length | 98 |
| Max. P | 0.585435 |

| Location | 1,931,455 – 1,931,553 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Shannon entropy | 0.45646 |
| G+C content | 0.43540 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.67 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1931455 98 + 22422827 ---UCCCCUCGUGGGUCUUUGGCAAA----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAU--- ---.....(((((.((..((((.(((----.((....)).......(((((.((((.....)))).)))))............))).)))).))...)))))...--- ( -18.00, z-score = -1.64, R) >droSim1.chrX 1374578 99 + 17042790 --CCCCCCUCGUGGGUUUUCGGCAAA----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAU--- --......(((((((((((((((...----.)))............(((((.((((.....)))).)))))....)))))))...............)))))...--- ( -20.56, z-score = -2.44, R) >droSec1.super_10 1716388 99 + 3036183 --CCCCCCUCGUGGGUUUUCGGCAAA----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAU--- --......(((((((((((((((...----.)))............(((((.((((.....)))).)))))....)))))))...............)))))...--- ( -20.56, z-score = -2.44, R) >droYak2.chrX 15375799 101 - 21770863 -CCCCCCCUCGUGGGUCUUCGGAAAA----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAAU-- -.......(((((.((.((.((((((----.((....)).......(((((.((((.....)))).)))))............)))))))).))...)))))....-- ( -19.70, z-score = -2.37, R) >droEre2.scaffold_4644 1903772 102 + 2521924 CCCCCCCCUCGUGGGUCUUCGGAAAA----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAAU-- ........(((((.((.((.((((((----.((....)).......(((((.((((.....)))).)))))............)))))))).))...)))))....-- ( -19.70, z-score = -2.27, R) >droAna3.scaffold_12929 1926012 83 + 3277472 ---------------UUUUUGAAAAGUCUCCGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACAC---------- ---------------((((((((........((....)).......(((((.((((.....)))).))))).))))))))..................---------- ( -11.60, z-score = -2.31, R) >droWil1.scaffold_181096 5452800 85 - 12416693 ------------------UCGUCGU-----CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGGAAACUCUUUUCCAAUACAUAUGUAUGUACAU ------------------.......-----.......(((((....(((((.((((.....)))).)))))....(((((....)))))......)))))........ ( -14.00, z-score = -1.44, R) >droVir3.scaffold_12928 7160911 105 + 7717345 ---GUUCGGGGUCGGACUGUCGCAUCAUACCGCCCCCGCAUGGCUACAAAAAUAGCAAAUUGUUAAUUUCGCUUCGGAAACUCUUUUCCAAUACACACACACACACAC ---...(((((.(((..((......))..))).)))))....((((......))))...................(((((....)))))................... ( -21.20, z-score = -1.11, R) >droMoj3.scaffold_6473 14030797 105 - 16943266 ---GUUCGGGGUCGGACUGUCGCAUCAUACCGCCCCCGCACAGCUACAAAAAUAGCAAAUUGUUAAUUUCGCUUCGGAAACUCUUUUCCAAUACACAAACACACACAC ---((.(((((.(((..((......))..))).))))).)).((((......))))...................(((((....)))))................... ( -23.70, z-score = -2.77, R) >consensus ___CCCCCUCGUGGGUCUUCGGCAAA____CGCCCCCGCAUAACUACGAAAAUAGCAAAUUGUUAAUUUCGCUUCGAAAACUCUUUUCCAAUACACACACGAAAA___ .....(((....)))..(((((.........((....)).......(((((.((((.....)))).)))))..))))).............................. ( -7.39 = -7.67 + 0.28)

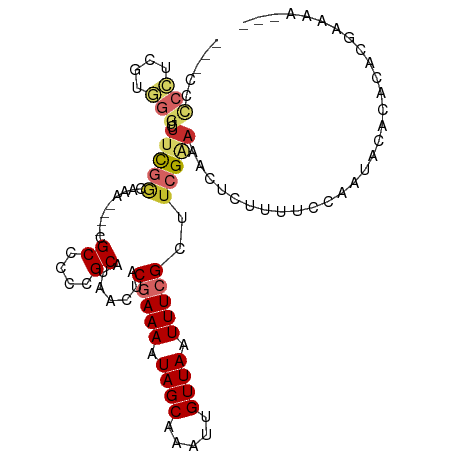
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:01 2011