| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,927,814 – 1,927,950 |
| Length | 136 |
| Max. P | 0.900816 |

| Location | 1,927,814 – 1,927,950 |
|---|---|
| Length | 136 |
| Sequences | 6 |
| Columns | 145 |
| Reading direction | reverse |
| Mean pairwise identity | 75.14 |
| Shannon entropy | 0.46383 |
| G+C content | 0.45150 |
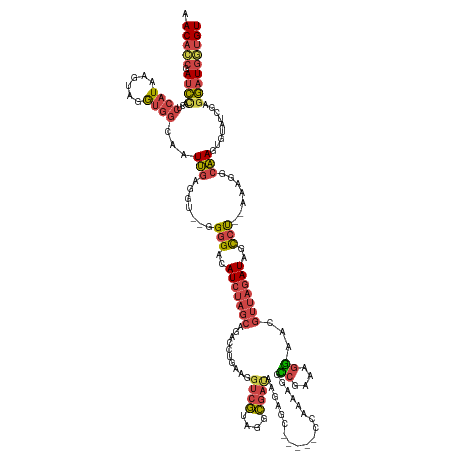
| Mean single sequence MFE | -38.51 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.77 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
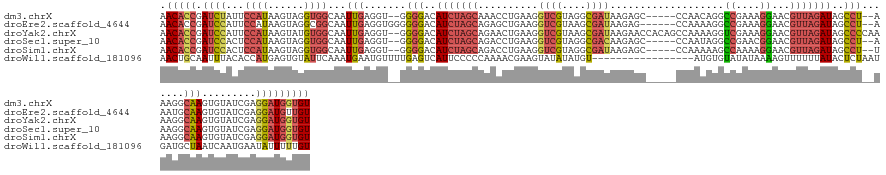
>dm3.chrX 1927814 136 - 22422827 AACACCGAUCUAUUCCAUAAGUAGGUGGCAAUUGAGGU--GGGGACAUCUAGCAAACCUGAAGGUCGUAGGCGAUAAGAGC-----CCAACAGGCCGAAAGGAACGUUAGAUAGCCU--AAAGGCAAGUGUAUCGAGGAUGGUGU .(((((.((((.((((.....(((((.((.....((((--(....))))).))..)))))..((((((.(((.......))-----)..)).))))....))))((.((.((.(((.--...)))..)).)).)).))))))))) ( -42.70, z-score = -2.23, R) >droEre2.scaffold_4644 1900078 137 - 2521924 AACACCGAUCCAUUCCAUAAGUAGGCGGCAAUUGAGGUGGGGGGACAUCUAGCAGAGCUGAAGGUCGUAAGCGAUAAGAG------CCAAAAGGCCGAAAGGAACGUUAGAUAGCCU--AAAUGCAAGUGUAUCGAGGAUGUUGU ..(..((((.((((.(((...(((((..............(....)(((((((.(.(((....((((....))))...))------))......((....))...))))))).))))--).)))..)))).))))..)....... ( -36.40, z-score = -0.77, R) >droYak2.chrX 15372176 143 + 21770863 AACACCGAUCCAUUCCAUAAGUAUGUGGCAAUUGAGGU--GGGGACAUCUAGCAGAACUGAAGGUCGUAAGCGAUAAGAACCACAGCCAAAAGGUCGAAAGGAACGUUAGAUAGCCCCAAAAGGCAAGUGUAUCGAGGAUGGUGU .(((((.((((....((((....))))(((.(((...(--((((..(((((((....((....((((....)))).))..((.(.(((....))).)...))...)))))))..))))).....))).))).....))))))))) ( -43.00, z-score = -2.77, R) >droSec1.super_10 1712792 136 - 3036183 AACACCGAUCCACUCCAUAAGUAGGUGGCAAUUGAGGU--GGGGACAUCUAGCAGACCUGAAGGUCGUAGGCGACAAGAGC-----CCAAUAGGCCGAACGGAACGUUAGAUAGCCU--AAAGGCAAGUGUAUCGAGGAUGGUGU .(((((.((((..(((.....(((((.((.....((((--(....))))).))..)))))..((((...(((.......))-----).....))))....))).((.((.((.(((.--...)))..)).)).)).))))))))) ( -45.40, z-score = -2.18, R) >droSim1.chrX 1370937 136 - 17042790 AACACCGAUCCACUCCAUAAGUAGGUGGCAAUUGAGGU--GGGGACAUCUAGCAGACCUGAAGGUCGUAGGCGAUAAGAGC-----CCAAAAAGCCAAAAGGAACGUUAGAUAGCCU--UAAGGCAAGUGUAUCGAGGAUGGUGU .(((((.((((.(.((.......)).)(((.(((...(--((((..(((((((.((((....))))((.(((.......))-----).......((....)).)))))))))..)))--))...))).))).....))))))))) ( -43.10, z-score = -2.43, R) >droWil1.scaffold_181096 5448524 128 + 12416693 AACUGCAAUUUACACCAUGAGUGUAUUCAAAUGAAUGUUUUGAGUCAUUCCCCCAAAACGAAGUAUAUAUGU-----------------AUGUGUAUAUAAAAGUUUUUUAUACUCUAAUGAUGCUAAUCAAUGAAUAUUUUUGU ....((((.......((((((((((.......(((((........)))))....(((((...((((((((..-----------------..))))))))....))))).)))))))...((((....))))))).......)))) ( -20.44, z-score = -0.74, R) >consensus AACACCGAUCCACUCCAUAAGUAGGUGGCAAUUGAGGU__GGGGACAUCUAGCAGACCUGAAGGUCGUAGGCGAUAAGAGC_____CCAAAAGGCCGAAAGGAACGUUAGAUAGCCU__AAAGGCAAGUGUAUCGAGGAUGGUGU .(((((.((((...((((......))))...(((.......(((..(((((((..........((((....))))...................((....))...)))))))..))).......))).........))))))))) (-20.65 = -21.77 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:08:00 2011