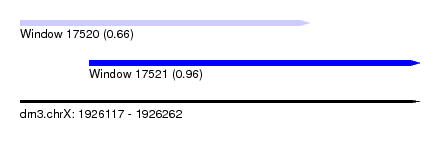
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,926,117 – 1,926,262 |
| Length | 145 |
| Max. P | 0.959757 |

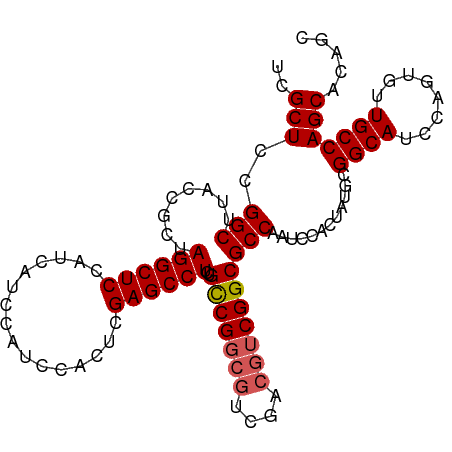
| Location | 1,926,117 – 1,926,222 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.57 |
| Shannon entropy | 0.09793 |
| G+C content | 0.63584 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -32.49 |
| Energy contribution | -32.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1926117 105 + 22422827 UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGUGGCAUCCAGUGUUGCCAGCACAGC ..(((..(((....((((((((((................))))))..(((((((....)))))))((((..........))))....))))..))))))..... ( -35.19, z-score = -1.48, R) >droSim1.chrX 1369534 105 + 17042790 UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGC ..(((..(((........((((((................))))))..(((((((....)))))))))).........(((((((........)))).))).))) ( -35.89, z-score = -1.63, R) >droSec1.super_10 1711106 105 + 3036183 UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGC ..(((..(((........((((((................))))))..(((((((....)))))))))).........(((((((........)))).))).))) ( -35.89, z-score = -1.63, R) >droYak2.chrX 15370543 99 - 21770863 UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAUCCACUCGAGCCUCUGCCG------ACGUCGGCGCCGAUCCACAAUGCGGCAUCCAGUGUUGCCAGCAUCGC ..((..((((........((((((................))))))..((((------....)))))))).......((((((((........)))).)))).)) ( -31.69, z-score = -1.03, R) >droEre2.scaffold_4644 1898385 105 + 2521924 UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAGCCACUCGAGCCUCUGUCGGCGUUGACGUCGGCGCCGAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGC ..(((((((....)))...((((.........))))....))))....((((((((((....))))))))))...((.(((((((........)))).))).)). ( -37.50, z-score = -1.11, R) >consensus UCGCUCCGGCUUACCGCUAGGCUCCAUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGC ..(((..(((........((((((................))))))..(((((((....))))))))))............((((........)))))))..... (-32.49 = -32.93 + 0.44)

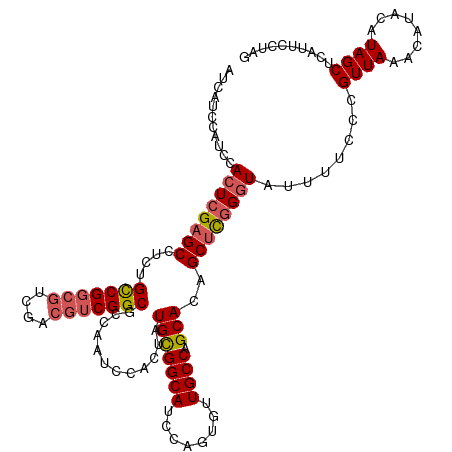
| Location | 1,926,142 – 1,926,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Shannon entropy | 0.19566 |
| G+C content | 0.54616 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -29.18 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1926142 120 + 22422827 AUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGUGGCAUCCAGUGUUGCCAGCACAGCUUGGGUAUUUUCCCGUUAAACAUACAUAGCUCAUUCCUAG ...........((((((((...(((.(((((((....)))))))...........(((((........))))))))..))))))))........((((........)))).......... ( -34.90, z-score = -2.45, R) >droSim1.chrX 1369559 120 + 17042790 AUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGCUCGGGUAUUUUCCCGUUAAAAAUACAUAGCUCAUUCUUAG ...........((((((((...(((.(((((((....)))))))............((((........)))).)))..))))))))........((((........)))).......... ( -36.40, z-score = -3.18, R) >droSec1.super_10 1711131 120 + 3036183 AUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGCUCGGGUAUUUUCCCGUUAAAAAUACAUAGCUCAUUCUUAG ...........((((((((...(((.(((((((....)))))))............((((........)))).)))..))))))))........((((........)))).......... ( -36.40, z-score = -3.18, R) >droYak2.chrX 15370568 110 - 21770863 AUCAUCCAUCCACUCGAGCCUCUGCCG------ACGUCGGCGCCGAUCCACAAUGCGGCAUCCAGUGUUGCCAGCAUCGCUCUGGUUUUUUCCCGUUAA-CAU---UAGCCCGUUUCUAG ...........(((.((((....((((------....))))...........((((((((........)))).)))).)))).)))........((((.-...---)))).......... ( -24.40, z-score = -0.29, R) >droEre2.scaffold_4644 1898410 118 + 2521924 AUCAUCCAGCCACUCGAGCCUCUGUCGGCGUUGACGUCGGCGCCGAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGCCCGGCU--UUUCCCGUUAAACAUACAUAGCUCGUUUCUCG ..............(((((....((((((((((....))))))))))...((.(((((((........)))).))).))..(((..--....))).............)))))....... ( -34.60, z-score = -1.50, R) >consensus AUCAUCCAUCCACUCGAGCCUCUGCCGGCGUCGACGUCGGCGCCAAUCCACUAUGCGGCAUCCAGUGUUGCCAGCACAGCUCGGGUAUUUUCCCGUUAAACAUACAUAGCUCAUUCCUAG ...........((((((((....(((((((....)))))))............(((((((........)))).)))..))))))))........((((........)))).......... (-29.18 = -29.90 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:59 2011