| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,835,366 – 1,835,480 |
| Length | 114 |
| Max. P | 0.719464 |

| Location | 1,835,366 – 1,835,480 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.36 |
| Shannon entropy | 0.68857 |
| G+C content | 0.49536 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.66 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
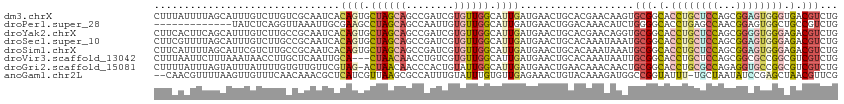
>dm3.chrX 1835366 114 - 22422827 CUUUAUUUUAGCAUUUGUCUUGUCGCAAUCACAGUGCUAGCAGCCGAUCGUGUUGGCAUUGAUGAACUGCACGAACAAGUGCGGCACCUGCUCCAGCGGAGUGGGUGACGUCUG ..........((((((((.((((.(((.((((((((((((((........))))))))))).)))..)))))))))))))))(.((((..((((...))))..)))).)..... ( -47.80, z-score = -3.60, R) >droPer1.super_28 920970 101 - 1111753 -------------UAUCUCAGGUUAAAUUGCGAAGCCUAGCAGCCAAUUGUGUUGGCAUUGAUGAACUGGACAAACAUCUGGGGCACCUGAGCCAACGGAGUGGCUGCCGUCUG -------------......(((((.........))))).(((((((.((.(((((((...(.((..(..((......))..)..)).)...))))))).)))))))))...... ( -35.30, z-score = -1.69, R) >droYak2.chrX 15303104 114 + 21770863 CUUCACUUCAGCAUUUGUCUUGCCGCAAUCACAGUGCUAGCAGCCGAUCGUGUUGGCAUUGAUGAACUGCACGAACAGGUGCGGCACCUGCUCCAGCGGGGUGGGAGACGUCUG ........(((....((((((((((((.((((((((((((((........))))))))))).))).(((......))).)))))))((..((((...))))..))))))).))) ( -45.50, z-score = -1.77, R) >droSec1.super_10 1628474 114 - 3036183 CUUCGUUUUAGCAUUUGUCUUGCCGCAAUCACAGUGCUAGCAGCCGAUCGUGUUGGCAUUGAUGAACUGCACAAAUAAAUGCGGCACCUGCUCCAGCGGAGUGGGAGACGUCUG ...(((((..((((((((.(((..(((.((((((((((((((........))))))))))).)))..))).)))))))))))....((..((((...))))..))))))).... ( -42.30, z-score = -2.24, R) >droSim1.chrX 1288628 114 - 17042790 CUUCAUUUUAGCAUUCGUCUUGCCGCAAUCACAGUGCUAGCAGCCGAUCGUGUUGGCAUUGAUGAACUGCACAAAUAAAUGCGGCACCUGCUCCAGCGGAGUGGGAGACGUCUG ...........((..((((((((((((....(((((((((((........))))))))))).((.....))........)))))))((..((((...))))..)))))))..)) ( -42.50, z-score = -2.50, R) >droVir3.scaffold_13042 1678018 111 - 5191987 CUUUAAUUCUUUAAAUAACCUUGCUCAAUUGCA---CUAACAACCUGUCGUGUUGGCAUUGAUGAACUGCACAAAUAAUUGCGGCACCUGCUCCAGCGGCGCCGGCGUCGUCUG .....................(((......)))---((((((........))))))....(((((...((.(((....)))((((.((.((....)))).)))))).))))).. ( -24.20, z-score = 0.16, R) >droGri2.scaffold_15081 2624561 113 - 4274704 CUUUUAUUUAGUAUUUAUUUUGUGUUGUUCGUAG-ACUAACAACCCACUGUAUUGGCAUUGAUGAACUGAACAAACAACUGCGGCACCUGCGCCAGAGGUGCCGGCGUCGUCUG ..((((((..((...(((..((.((((((.(...-.).)))))).))..)))...))...))))))..............(((((.((.(((((...))))).)).)))))... ( -27.50, z-score = -0.22, R) >anoGam1.chr2L 37213803 111 + 48795086 --CAACGUUUUAAGUUGUUUCAACAAACGCUCAUCGUUAAGCGCCAUUUGUAUUUGUGUUGAGAAACUGUACAAAGAUGGCCGGUAUUU-UGCUAAUAUCCGAGCUAACGUUCG --.((((((....((((...))))....((((........(.(((((((((((....(((....))).)))))..)))))))((((((.-....)))))).)))).)))))).. ( -26.50, z-score = -1.15, R) >consensus CUUCAUUUUAGCAUUUGUCUUGCCGCAAUCACAGUGCUAGCAGCCGAUCGUGUUGGCAUUGAUGAACUGCACAAACAAGUGCGGCACCUGCUCCAGCGGAGUGGGAGACGUCUG ...............................((((.((((((........)))))).))))...................(((.(.((((((((...)))))))).).)))... (-11.64 = -12.66 + 1.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:49 2011