| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,795,561 – 1,795,676 |
| Length | 115 |
| Max. P | 0.704833 |

| Location | 1,795,561 – 1,795,676 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Shannon entropy | 0.63663 |
| G+C content | 0.48663 |
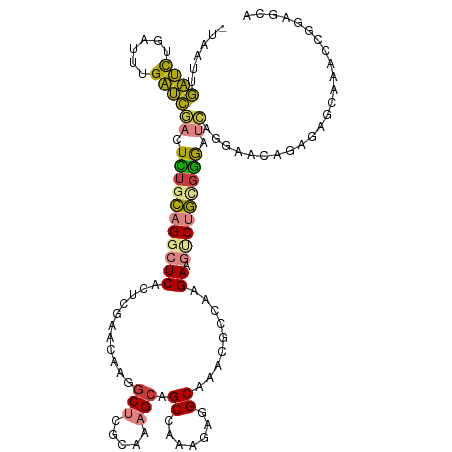
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -15.75 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.75 |
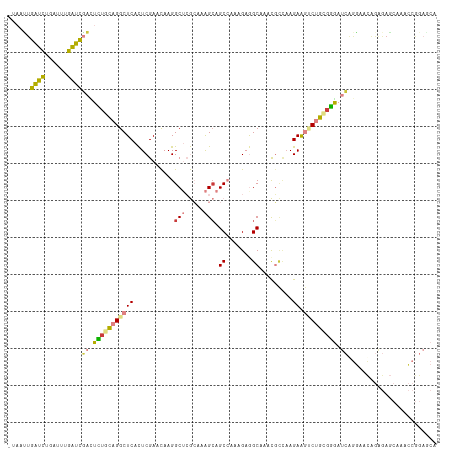
| Combinations/Pair | 1.50 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
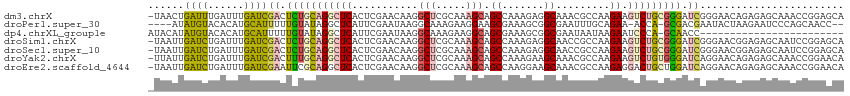
>dm3.chrX 1795561 115 - 22422827 -UAACUGAUUUGAUUUGAUCGACUCUGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAAGAGGCAAACGCCAAGAAGUCUGCGGGAUCGGGAACAGAGAGCAAACCGGAGCA -...(((.((((.((((.((((..((((((((((..........((((.((...)))))).....(((....)))..).)))))))))..))))...))))...)))).))).... ( -39.90, z-score = -2.56, R) >droPer1.super_30 96293 108 + 958394 ----AUAUGUACACAUGCAUUUUUGUAUAGGCUCAUUCGAAUAAGGCAAAGAAGGAAGCGAAAGCGGCGAAUUUGCAGAA-ACCA-GCGACGAAUACUAAGAAUCCCAGCAACC-- ----...........(((.(((((((......((....)).....))))))).(((.((....)).......((((....-....-)))).............)))..)))...-- ( -17.00, z-score = 0.91, R) >dp4.chrXL_group1e 358589 91 - 12523060 AUACAUAUGUACACAUGCAUUUUUGUAUAGGCUCAUUCGAAUAAGGCAAAGAAGGCAGCGAAAGCGGCGAAUAAUAAGAAUCCCA-GCAACC------------------------ (((((.(((((....)))))...)))))..(((.((((.......((.......)).((....))............))))...)-))....------------------------ ( -15.70, z-score = -0.16, R) >droSim1.chrX 1267958 115 - 17042790 -UAAUUGAUCUGAUUUGAUCGACUCUGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAAGAGGCAACCGCCAAGAAGUCUGCGGGAUCGGGAACGGAGAGCAAUCCGGAGCA -.....((((......))))((..((((((((((..........((((.((...)))))).....(((....)))..).)))))))))..)).....((((......))))..... ( -39.40, z-score = -1.55, R) >droSec1.super_10 1596117 115 - 3036183 -UAAUUGAUCUGAUUUGAUCGACUCUGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAAGAGGCAACCGCCAAGAAGUCUGCGGGAUCGGGAACGGAGAGCAAUCCGGAGCA -.....((((......))))((..((((((((((..........((((.((...)))))).....(((....)))..).)))))))))..)).....((((......))))..... ( -39.40, z-score = -1.55, R) >droYak2.chrX 15270016 115 + 21770863 -UUAUUGAUCUGAUUUGAUCGACUUUGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAAGAAGCAAACGCCAAGAAGUCUGUGGGAUCAGGAACAGAGAGCAAACCGGAACA -.......((((((((.((.((((((...(((((....))....((((.((...))))))............)))..)))))).)).))))))))..................... ( -28.30, z-score = -0.02, R) >droEre2.scaffold_4644 1774495 115 - 2521924 -UAAUUGAUCUGAUUUGAUCGAAUUCGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAGGAAGCAAACGCCAAGAGGACUGCUGGAUCAGGAACAGAGAGCAAACCGGAACA -...(((.((((.((((((((....)((((.(((..........((((.((...))))))......((....))...)))..))))..)))))))..))))...)))......... ( -27.60, z-score = 0.04, R) >consensus _UAAUUGAUCUGAUUUGAUCGACUCUGCAGGCUCACUCGAACAAGGCUCGCAAAGCAGCCAAAGAGGCAAACGCCAAGAAGUCUGCGGGAUCAGGAACAGAGAGCAAACCGGAGCA ......((((......))))((.(((((((((((...........(((.....))).((.......)).........)).))))))))).))........................ (-15.75 = -15.00 + -0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:43 2011