
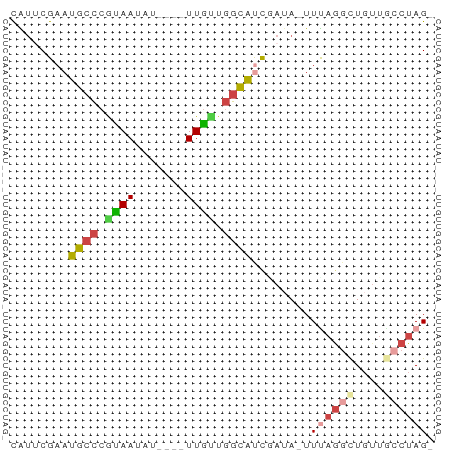
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,774,509 – 1,774,561 |
| Length | 52 |
| Max. P | 0.970141 |

| Location | 1,774,509 – 1,774,561 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.49031 |
| G+C content | 0.41645 |
| Mean single sequence MFE | -11.97 |
| Consensus MFE | -5.65 |
| Energy contribution | -6.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1774509 52 + 22422827 CAUGCGAAUGCCCGUAAUAU----UUGCUGGCAUCAAUA-UUUAAUCUGUUGCCUAG- ...(((((((.......)))----)))).((((.((...-.......)).))))...- ( -9.60, z-score = -0.66, R) >droSim1.chrX 1247631 52 + 17042790 CGUGCGAAUGCCCGUAAUAU----UUGUUGGCAUCGAUA-UUCAGGCUGUUGCCUAG- ....(((.((((..((....----.))..)))))))...-...((((....))))..- ( -12.90, z-score = -0.85, R) >droSec1.super_10 1575012 52 + 3036183 CGUUCGAAUGCCCGUAAUAU----UUGCUGGCAUCGAUA-UUCAGGCUGUUGCCUAG- ...((((.((((.(((....----.))).))))))))..-...((((....))))..- ( -17.10, z-score = -2.83, R) >droYak2.chrX 15252902 52 - 21770863 CAUUCGAAUGCCCACAAUGU----UUGUUGGCAUCCAUA-UUUAGGCUGUUACCUAG- .....(.(((((.(((....----.))).))))).)...-..((((......)))).- ( -11.60, z-score = -1.51, R) >droEre2.scaffold_4644 1753324 52 + 2521924 UGUUCGAAUGCCCAUAAUAU----UUGUUGGCACUCAAA-UUUAGGUUGUUACCUAG- .....((.((((.(((....----.))).)))).))...-..(((((....))))).- ( -12.10, z-score = -2.22, R) >droVir3.scaffold_12970 9884528 58 - 11907090 CAAUCGAACAAUCGUAACGUAAUCUUAAUUAUGCCGAUAGUUUUGGUUCAUCACUAGU .((((((((.((((...((((((....)))))).)))).).))))))).......... ( -8.50, z-score = -0.90, R) >consensus CAUUCGAAUGCCCGUAAUAU____UUGUUGGCAUCGAUA_UUUAGGCUGUUGCCUAG_ ........((((.((((.......)))).))))........((((((....)))))). ( -5.65 = -6.15 + 0.50)


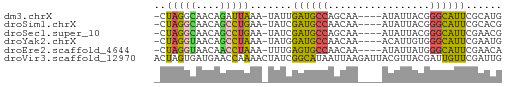
| Location | 1,774,509 – 1,774,561 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.49031 |
| G+C content | 0.41645 |
| Mean single sequence MFE | -11.94 |
| Consensus MFE | -7.00 |
| Energy contribution | -6.45 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1774509 52 - 22422827 -CUAGGCAACAGAUUAAA-UAUUGAUGCCAGCAA----AUAUUACGGGCAUUCGCAUG -...((((.(((......-..))).)))).....----.........((....))... ( -7.50, z-score = -0.01, R) >droSim1.chrX 1247631 52 - 17042790 -CUAGGCAACAGCCUGAA-UAUCGAUGCCAACAA----AUAUUACGGGCAUUCGCACG -.(((((....)))))..-...(((((((.....----........)))).))).... ( -12.12, z-score = -2.05, R) >droSec1.super_10 1575012 52 - 3036183 -CUAGGCAACAGCCUGAA-UAUCGAUGCCAGCAA----AUAUUACGGGCAUUCGAACG -.(((((....)))))..-..((((((((.....----........)))).))))... ( -14.62, z-score = -2.79, R) >droYak2.chrX 15252902 52 + 21770863 -CUAGGUAACAGCCUAAA-UAUGGAUGCCAACAA----ACAUUGUGGGCAUUCGAAUG -.(((((....)))))..-..((((((((.(((.----....))).)))))))).... ( -17.10, z-score = -3.64, R) >droEre2.scaffold_4644 1753324 52 - 2521924 -CUAGGUAACAACCUAAA-UUUGAGUGCCAACAA----AUAUUAUGGGCAUUCGAACA -.(((((....)))))..-((((((((((.....----........)))))))))).. ( -15.92, z-score = -4.17, R) >droVir3.scaffold_12970 9884528 58 + 11907090 ACUAGUGAUGAACCAAAACUAUCGGCAUAAUUAAGAUUACGUUACGAUUGUUCGAUUG ........(((((.......((((((.((((....)))).))..)))).))))).... ( -4.40, z-score = 1.32, R) >consensus _CUAGGCAACAGCCUAAA_UAUCGAUGCCAACAA____AUAUUACGGGCAUUCGAACG ..(((((....))))).......((((((.................))))))...... ( -7.00 = -6.45 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:40 2011