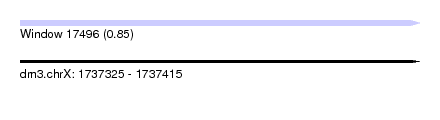
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,737,325 – 1,737,415 |
| Length | 90 |
| Max. P | 0.851361 |

| Location | 1,737,325 – 1,737,415 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.41 |
| Shannon entropy | 0.58750 |
| G+C content | 0.49247 |
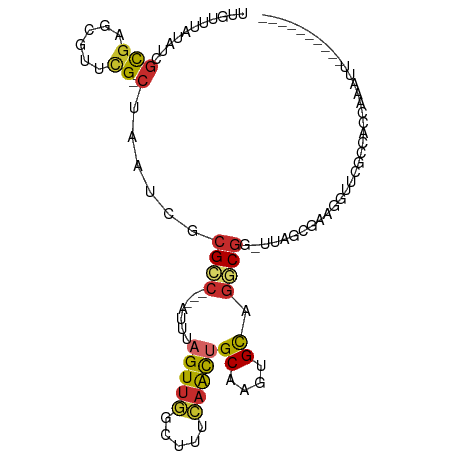
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -6.63 |
| Energy contribution | -6.51 |
| Covariance contribution | -0.12 |
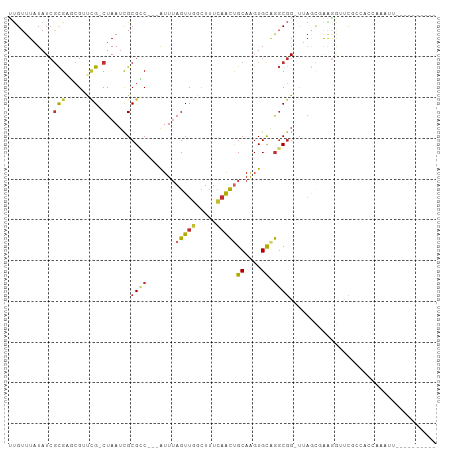
| Combinations/Pair | 1.62 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
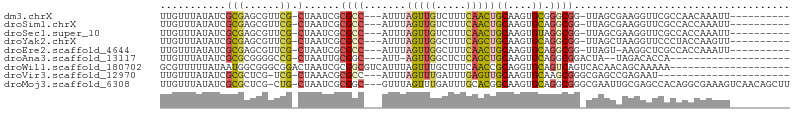
>dm3.chrX 1737325 90 + 22422827 UUGUUUAUAUCGCGAGCGUUCG-CUAAUCGCGCC---AUUUAGUUGUCUUUCAACUGCAAGUGCGGGCGG-UUAGCGAAGGUUCGCCAACAAAUU---------- ...........((((((.((((-((((((((.((---(((((((((.....))))))..)))).).))))-)))))))).)))))).........---------- ( -41.20, z-score = -5.94, R) >droSim1.chrX 1229897 90 + 17042790 UUGUUUAUAUCGCGAGCGUUCG-CUAAUCGCGCC---AUUUAGUUGUCUUUCAACUGCAAGUGCAGGCGG-UUAGCGAAGGUUCGCCACCAAAUU---------- ...........((((((.((((-((((((((..(---(((((((((.....))))))..))))...))))-)))))))).)))))).........---------- ( -37.70, z-score = -5.19, R) >droSec1.super_10 1538255 90 + 3036183 UUGUUUAUAUCGCGAGCGUUCG-CUAAUCGCGCC---AUUUAGUUGUCUUUCAACUGCAAGUGUAGGCGG-UUAGCGAAGGUUCGCCACCAAAUU---------- ...........((((((.((((-((((((((..(---(((((((((.....))))))..))))...))))-)))))))).)))))).........---------- ( -37.70, z-score = -5.51, R) >droYak2.chrX 1582244 90 + 21770863 UUGUUUAUAUCGCGAGCGUUCG-CUAAUCGCGCC---AUUUAGUUGGCUUUCAGCUGCAAGUGCAGGCGG-UUAGCUAAGGUUCCCUACCAAGUU---------- .............((((.((.(-(((((((((((---(......))))......((((....))))))))-))))).)).))))...........---------- ( -32.20, z-score = -2.75, R) >droEre2.scaffold_4644 1716596 89 + 2521924 UUGUUUAUAUCGCGAGCGUUCG-CUAAUCGCGCC---AUUUAGUUGGCUUUCAACUGCAAGUGCAGGCGG-UUAGU-AAGGCUCGCCACCAAAUU---------- ...........((((((.((.(-(((((((((((---(......))))......((((....))))))))-)))))-)).)))))).........---------- ( -38.10, z-score = -5.05, R) >droAna3.scaffold_13117 1039028 78 + 5790199 UUGUUUAUAUCGCGCGGGGCCG-CUAAUUGCGGC---AUU-AGUUGGCUCUCAGCUGCAAGUGCAGGCGGACUA--UAGACACCA-------------------- .((((((((((.((((((((((-(((((......---)))-)).)))))))...((((....))))))))).))--))))))...-------------------- ( -30.00, z-score = -1.96, R) >droWil1.scaffold_180702 3107060 85 + 4511350 GCGUUUUUAUAAUGGCGGGCGGACUAAUCGCGGCGUCAUUUAGUUUGCUUUCAACCGCAGGUGCAGUCAGUCACAACAGCAAAAA-------------------- ((..........(((((((((((((((............)))))))))))......((....)).)))).........)).....-------------------- ( -17.91, z-score = 0.81, R) >droVir3.scaffold_12970 10715797 78 - 11907090 UUGUUUAUAUCGCGCUCG-UCG-CUAAACGCGCC---AUUUAGUUUGAUUUGAGUUGCAAGUGCAAGCGGGCGAGCCGAGAAU---------------------- .........(((.(((((-(((-((..((..((.---((((((......)))))).))..))...))).))))))))))....---------------------- ( -25.20, z-score = -0.96, R) >droMoj3.scaffold_6308 1202997 100 + 3356042 UUGUUUAUAUCGCGCUCG-CUG-CUAAUCGCGGC---GUUUAGUUUGAUUUGCACGGCAAGUGCAGGCGGGCGAAUUGCGAGCCACAGGCGAAAGUCAACAGCUU ...........(((((((-(((-(.....)))))---((...(((((.(((((((.....)))))))))))).....))))))....(((....)))....)).. ( -35.60, z-score = -0.59, R) >consensus UUGUUUAUAUCGCGAGCGUUCG_CUAAUCGCGCC___AUUUAGUUGGCUUUCAACUGCAAGUGCAGGCGG_UUAGCGAAGGUUCGCCACCAAAUU__________ ..........((((..............))))......................((((....))))....................................... ( -6.63 = -6.51 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:37 2011