| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,730,410 – 1,730,510 |
| Length | 100 |
| Max. P | 0.996174 |

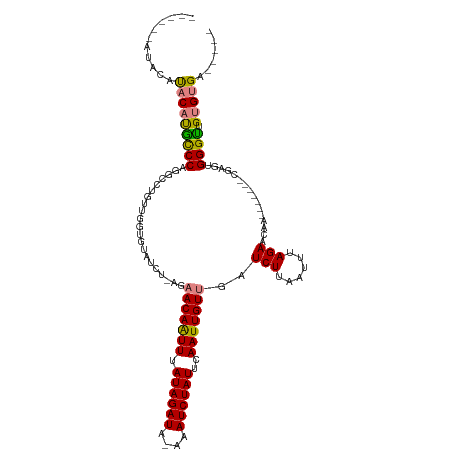
| Location | 1,730,410 – 1,730,510 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.38360 |
| G+C content | 0.36726 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1730410 100 + 22422827 ------AUACAUACAUGCCCAGGCCUGUUGGUGUAUCU-AGAACAAUUUAUAGAUA-AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA-------CGAGUGGGUUGUGUGA----- ------...((((((.(((((.(((....))).(((((-(..........))))))-............((((((.(((......))).)))-------))).))))))))))).----- ( -27.30, z-score = -2.64, R) >droSim1.chrX 1222914 100 + 17042790 ------AUACAUACAUGCCCAGGUCUGUUGGUGUAUCU-AGAACAAUUUAUAGAUA-AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA-------CGAGUGGGUUGUGUGA----- ------...((((((.(((((.((((((...(((....-...)))....)))))).-............((((((.(((......))).)))-------))).))))))))))).----- ( -27.10, z-score = -2.59, R) >droSec1.super_10 1531289 100 + 3036183 ------AUACAUACAUGCCCAGGUCUGUUGGUGUAUCU-AGAACAAUUUAUAGAUA-AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA-------CGAGUGGGUUGUGUGA----- ------...((((((.(((((.((((((...(((....-...)))....)))))).-............((((((.(((......))).)))-------))).))))))))))).----- ( -27.10, z-score = -2.59, R) >droYak2.chrX 1574913 96 + 21770863 ----------AUACACACCCAGGCCUGUUGGUUUAUCU-AGCACAAUUUAUAGAUA-AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA-------CGAGUGGGUUGUGUGA----- ----------.((((((((((.(((....)))((((((-(..........))))))-)...........((((((.(((......))).)))-------))).)))).)))))).----- ( -26.50, z-score = -2.85, R) >droEre2.scaffold_4644 1709500 96 + 2521924 ----------AUACAUACCCAGGCCUGUUGGUGUAUCU-AGCACAAUUUAUAGAUA-AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA-------CGAGUGGGUUGUGUGA----- ----------.((((((((((.(((....))).(((((-(..........))))))-............((((((.(((......))).)))-------))).)))).)))))).----- ( -24.30, z-score = -1.91, R) >dp4.chrXL_group1a 6608367 120 + 9151740 ACACACACACUCUCUCGCCCACACUCGAGUGUGUGUUUUAGAACAGUUUAUAGAUACAAAUCUAUUCAAUUGUUGAUCUUUAUUUAGAUUGGUGGGGGGCGGGAGGGCUGUGUGUGGGUC .(((((((((((((((((((.((((................(((((((.((((((....))))))..)))))))(((((......)))))))))..))))))))))).)))))))).... ( -51.60, z-score = -6.20, R) >droPer1.super_25 744298 120 - 1448063 AUACACACACUCUCUCGUCCACACUCGAGUGUGUGUUUUAGAACAGUUUAUAGAUACAAAUCUAUUCAAUUGUUGAUCUUUAUUUAGAAUGGUGGGGGGCGGGAGGGCUGUGUGUGGGUC .(((((((((((((((((((.((((................(((((((.((((((....))))))..)))))))..(((......)))..))))..))))))))))).)))))))).... ( -44.40, z-score = -4.60, R) >consensus ______AUACAUACAUGCCCAGGCCUGUUGGUGUAUCU_AGAACAAUUUAUAGAUA_AAAUCUAUUCAAUUGUUGAUCUUAAUUUAGAACAA_______CGAGUGGGUUGUGUGA_____ ...........(((((((((.....................(((((((.((((((....))))))..)))))))..(((......)))................)))).)))))...... (-17.39 = -17.07 + -0.32)

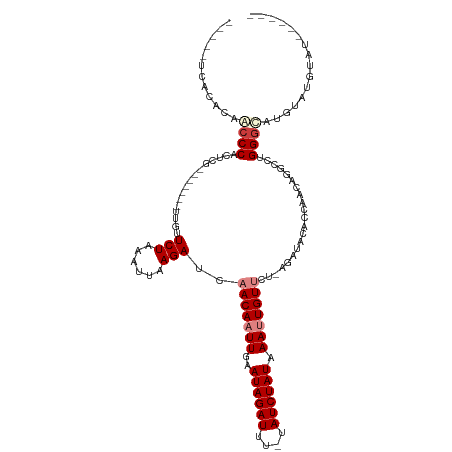
| Location | 1,730,410 – 1,730,510 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.38360 |
| G+C content | 0.36726 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -11.36 |
| Energy contribution | -12.34 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1730410 100 - 22422827 -----UCACACAACCCACUCG-------UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU-UAUCUAUAAAUUGUUCU-AGAUACACCAACAGGCCUGGGCAUGUAUGUAU------ -----.((.(((.(((((..(-------(((.(((......)))..(((((((..((((((..-.)))))).)))))))..-........))))..)..))))..))).))...------ ( -17.70, z-score = -0.93, R) >droSim1.chrX 1222914 100 - 17042790 -----UCACACAACCCACUCG-------UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU-UAUCUAUAAAUUGUUCU-AGAUACACCAACAGACCUGGGCAUGUAUGUAU------ -----.((.(((.((((....-------(((((....(((.(((...((((((..((((((..-.)))))).)))))))))-.))).....)))))...))))..))).))...------ ( -17.20, z-score = -1.04, R) >droSec1.super_10 1531289 100 - 3036183 -----UCACACAACCCACUCG-------UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU-UAUCUAUAAAUUGUUCU-AGAUACACCAACAGACCUGGGCAUGUAUGUAU------ -----.((.(((.((((....-------(((((....(((.(((...((((((..((((((..-.)))))).)))))))))-.))).....)))))...))))..))).))...------ ( -17.20, z-score = -1.04, R) >droYak2.chrX 1574913 96 - 21770863 -----UCACACAACCCACUCG-------UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU-UAUCUAUAAAUUGUGCU-AGAUAAACCAACAGGCCUGGGUGUGUAU---------- -----..(((((.(((((..(-------(((......(((.((....((((((..((((((..-.)))))).)))))).))-.)))....))))..)..)))))))))..---------- ( -22.50, z-score = -2.46, R) >droEre2.scaffold_4644 1709500 96 - 2521924 -----UCACACAACCCACUCG-------UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU-UAUCUAUAAAUUGUGCU-AGAUACACCAACAGGCCUGGGUAUGUAU---------- -----....(((((((((..(-------(((......(((.((....((((((..((((((..-.)))))).)))))).))-.)))....))))..)..))))).)))..---------- ( -19.10, z-score = -1.53, R) >dp4.chrXL_group1a 6608367 120 - 9151740 GACCCACACACAGCCCUCCCGCCCCCCACCAAUCUAAAUAAAGAUCAACAAUUGAAUAGAUUUGUAUCUAUAAACUGUUCUAAAACACACACUCGAGUGUGGGCGAGAGAGUGUGUGUGU ....((((((((.(.(((.(((((..((((.((((......))))........((((((.(((((....)))))))))))..............).))).))))).))).))))))))). ( -36.70, z-score = -4.96, R) >droPer1.super_25 744298 120 + 1448063 GACCCACACACAGCCCUCCCGCCCCCCACCAUUCUAAAUAAAGAUCAACAAUUGAAUAGAUUUGUAUCUAUAAACUGUUCUAAAACACACACUCGAGUGUGGACGAGAGAGUGUGUGUAU .....(((((((.(.(((.((................................((((((.(((((....))))))))))).......(((((....)))))..)).))).)))))))).. ( -27.40, z-score = -3.01, R) >consensus _____UCACACAACCCACUCG_______UUGUUCUAAAUUAAGAUCAACAAUUGAAUAGAUUU_UAUCUAUAAAUUGUUCU_AGAUACACCAACAGGCCUGGGCAUGUAUGUAU______ ............((((................(((......)))..(((((((..((((((....)))))).))))))).....................))))................ (-11.36 = -12.34 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:36 2011