| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,726,029 – 1,726,130 |
| Length | 101 |
| Max. P | 0.989816 |

| Location | 1,726,029 – 1,726,130 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.51983 |
| G+C content | 0.55630 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
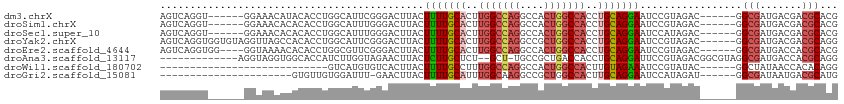
>dm3.chrX 1726029 101 - 22422827 AGUCAGGU------GGAAACAUACACCUGGCAUUCGGGACUUACUUUUGCACUUGGCCAGGCCACUGGCCACCUGCAGGAAUCCGUAGAC------GGCGAUGACGACGCACG .(((((((------(........)))))))).....(((.....(((((((..(((((((....)))))))..))))))).))).....(------((((.......))).)) ( -40.80, z-score = -3.71, R) >droSim1.chrX 1218549 101 - 17042790 AGUCAGGU------GGAAACACACACCUGGCAUUUGGGACUUACUUUUGCACUUGGCCAGGCCACUGGCCACCUGCAGGAAUCCGUAGAC------GGCGAUGACGACGCACG .(((((((------(........)))))))).....(((.....(((((((..(((((((....)))))))..))))))).))).....(------((((.......))).)) ( -40.80, z-score = -3.72, R) >droSec1.super_10 1526974 101 - 3036183 AGUCAGGU------GGAAACACACACCUGGCAUUUGGGACUUACUUUUGCACUUGGCCAGGCCACUGGCCACCUGCAGGAAUCCAUAGAC------GGCGAUGACGACGCACG .(((((((------(........)))))))).....(((.....(((((((..(((((((....)))))))..))))))).))).....(------((((.......))).)) ( -40.80, z-score = -4.33, R) >droYak2.chrX 1570384 107 - 21770863 AGUCAGGUGGUGUAGGUUAGCCACACCUGGCAUUCGGGACUUACUUUUGCACUUGGCCAGGCCGCUGGCCACCUGCAGGAAUCCGUAGAC------GGCGAUGACGACGCAGG .((((((((..((......))..))))))))..(((........(((((((..(((((((....)))))))..)))))))..(((....)------))......)))...... ( -39.90, z-score = -0.68, R) >droEre2.scaffold_4644 1705101 103 - 2521924 AGUCAGGUGG----GGUAAAACACACCUGGCGUUCGGGACUUACUUUUGCACUUGGCCAGGCCACUGGCCACCUGCAGGAAUCCGUAGAC------GGCGAUGACCACGCACG .((((((((.----(......).))))))))((.((((......(((((((..(((((((....)))))))..)))))))..(((....)------))......)).)).)). ( -43.40, z-score = -2.87, R) >droAna3.scaffold_13117 1031854 97 - 5790199 -------------AGGUAGGUGGCACCAUCUUGGUAGAACUUACUCUUGCUCU--GCU-UGCCGCUGACCACCUGCAGGAUUCCGUAGACGGCGUAGGCGAUGACCACGCAGG -------------.(((.(((((((.......((((((.(........).)))--)))-))))))).))).(((((......(((....))).((.((......))))))))) ( -33.11, z-score = -0.43, R) >droWil1.scaffold_180702 3094163 79 - 4511350 ----------------------------GUCAUGUGUCACUUACUUUUGCCUUUGGCCAGGCCACUGGCCACUUGUAGAAAUCCGUAUAC------GGCUAUAACCACACAGG ----------------------------....(((((.......((((((...(((((((....)))))))...))))))..(((....)------))........))))).. ( -24.10, z-score = -2.24, R) >droGri2.scaffold_15081 2470339 84 + 4274704 ----------------------GUGUUGUGGAUUU-GAACUUACUUUUGCAUUUGGCAAGGCCGCUGGCCACUUGCAGGAAUCCAUAGAU------GGCGAUAAUGACGCAUG ----------------------...((((((((((-..........(((((..((((.((....)).))))..)))))))))))))))..------.(((.......)))... ( -23.80, z-score = -0.87, R) >consensus AGUCAGGU______GGAAACACACACCUGGCAUUUGGGACUUACUUUUGCACUUGGCCAGGCCACUGGCCACCUGCAGGAAUCCGUAGAC______GGCGAUGACGACGCACG ............................................(((((((..(((((((....)))))))..))))))).................(((.......)))... (-18.43 = -19.26 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:34 2011