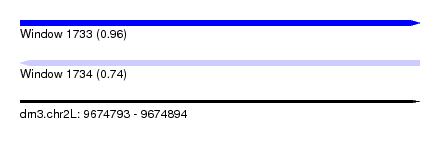
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,674,793 – 9,674,894 |
| Length | 101 |
| Max. P | 0.962332 |

| Location | 9,674,793 – 9,674,894 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.22 |
| Shannon entropy | 0.21821 |
| G+C content | 0.52149 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -20.97 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
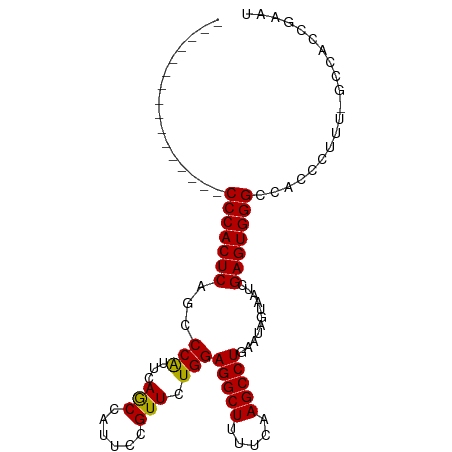
>dm3.chr2L 9674793 101 + 23011544 CAUUCAGACCACUCAGCCCACUCAGCCCGUUCAGCCAUUACGUUAUGGAGGCUUUUCAAGCCUGAAUAGUAAUCGAGUGGGCCACCCUUUUGCCAACGAAU .((((.((....)).((((((((.((..((((..((((......))))(((((.....))))))))).))....))))))))...............)))) ( -27.50, z-score = -2.31, R) >droSec1.super_3 5121213 84 + 7220098 ----------------CCCACUCAGCCCACUCAACCAUUCCGUUCUGGAGGCUUUUCAAGCCUGAAUAGUAAUCGAGUGGGCCACCCUUU-GCCACCGAAU ----------------........((((((((..(((........)))(((((.....)))))...........))))))))........-.......... ( -25.10, z-score = -2.82, R) >droSim1.chr2L 9452550 84 + 22036055 ----------------CCCACUCAACCCAUUCAGCCAUUCCGUUCUGGAGGCUUUUCAAGCCUGAAUAGUAAUCGAGUGGGCCACCCUUU-GCCACCGAAU ----------------(((((((...........(((........)))(((((.....)))))...........))))))).........-.......... ( -20.80, z-score = -1.34, R) >consensus ________________CCCACUCAGCCCAUUCAGCCAUUCCGUUCUGGAGGCUUUUCAAGCCUGAAUAGUAAUCGAGUGGGCCACCCUUU_GCCACCGAAU ................(((((((...(((...(((......))).)))(((((.....)))))...........))))))).................... (-20.97 = -20.53 + -0.44)

| Location | 9,674,793 – 9,674,894 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.22 |
| Shannon entropy | 0.21821 |
| G+C content | 0.52149 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
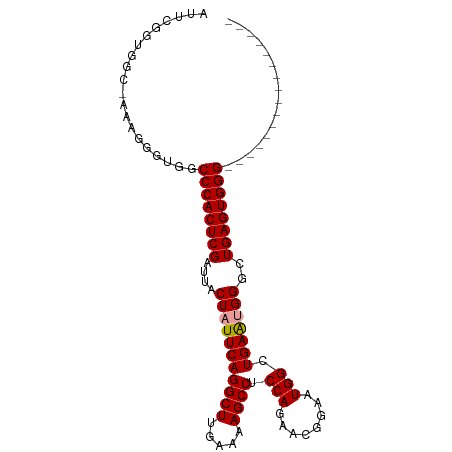
>dm3.chr2L 9674793 101 - 23011544 AUUCGUUGGCAAAAGGGUGGCCCACUCGAUUACUAUUCAGGCUUGAAAAGCCUCCAUAACGUAAUGGCUGAACGGGCUGAGUGGGCUGAGUGGUCUGAAUG (((((..(((....(..((((((((((((.......)).((((((...((((.............))))...))))))))))))))))..).)))))))). ( -33.82, z-score = -1.70, R) >droSec1.super_3 5121213 84 - 7220098 AUUCGGUGGC-AAAGGGUGGCCCACUCGAUUACUAUUCAGGCUUGAAAAGCCUCCAGAACGGAAUGGUUGAGUGGGCUGAGUGGG---------------- ((((....((-.....))(((((((((((((((..((((((((.....)))))...)))..)..))))))))))))))))))...---------------- ( -30.30, z-score = -1.74, R) >droSim1.chr2L 9452550 84 - 22036055 AUUCGGUGGC-AAAGGGUGGCCCACUCGAUUACUAUUCAGGCUUGAAAAGCCUCCAGAACGGAAUGGCUGAAUGGGUUGAGUGGG---------------- ....(((.((-.....)).)))((((((((..(((((((((((.....))))(((.....))).....)))))))))))))))..---------------- ( -29.10, z-score = -1.61, R) >consensus AUUCGGUGGC_AAAGGGUGGCCCACUCGAUUACUAUUCAGGCUUGAAAAGCCUCCAGAACGGAAUGGCUGAAUGGGCUGAGUGGG________________ ....................((((((((....(((((((((((.....)))).(((........))).)))))))..))))))))................ (-25.25 = -25.37 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:46 2011