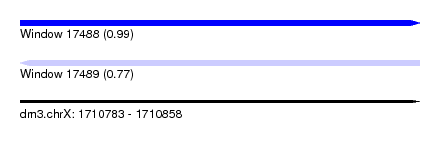
| Sequence ID | dm3.chrX |
|---|---|
| Location | 1,710,783 – 1,710,858 |
| Length | 75 |
| Max. P | 0.993439 |

| Location | 1,710,783 – 1,710,858 |
|---|---|
| Length | 75 |
| Sequences | 5 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.44658 |
| G+C content | 0.60368 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.64 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
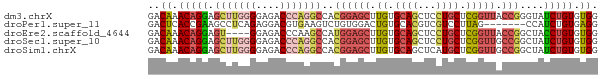
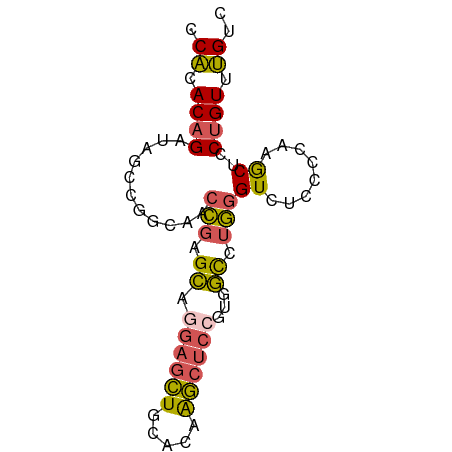
>dm3.chrX 1710783 75 + 22422827 GACAAACAGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGGUAUCUGUGUGG ..((.(((((.(((((((....)))))))...((((((.....))))))(((((((...)))))))))))).)). ( -37.90, z-score = -3.53, R) >droPer1.super_11 880756 68 + 2846995 GACUCACCGAAGCCUCAGAGGACGUGAAGUCUGUGGACUGUGCACGUCGUCCUUAG-------CCAUCUGUGAGG ..(((((.((.((....((.((((((.((((....))))...)))))).))....)-------)..)).))))). ( -27.20, z-score = -2.24, R) >droEre2.scaffold_4644 1691576 71 + 2521924 GACAAACAGGAGU----GGAGACCCAAGCCAUGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGCUACCUGUGUGG ..((.((((((((----((.((((..(((...((((((.....)))))).))).)))).)).))).))))).)). ( -30.00, z-score = -2.37, R) >droSec1.super_10 1505631 75 + 3036183 GACAAACAGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUGCCGGCUAUCUGUGUGG ..((.(((((.(((((((....)))))))...((((((.....)))))).((.(((...)))))..))))).)). ( -34.70, z-score = -2.13, R) >droSim1.chrX 1203918 75 + 17042790 GACAAACAGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCAUGCUCGGUUGCCGGCUAUCUGUGUGG ..((.(((((((((((((....))))(((.((.((((.((.......)).)))).)).))))))).))))).)). ( -35.10, z-score = -2.66, R) >consensus GACAAACAGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGCUAUCUGUGUGG ..((.(((((.(((((((....)))))))..(((...(((.((((...)))).)))...)))....))))).)). (-19.36 = -20.64 + 1.28)
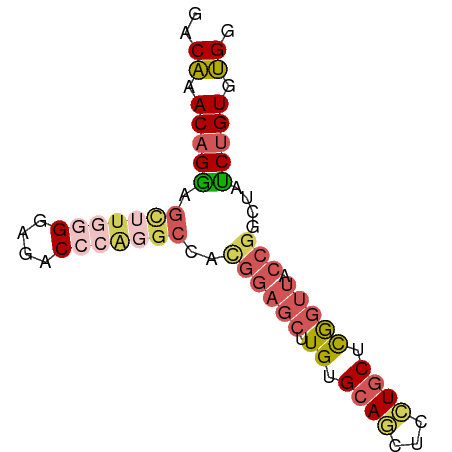
| Location | 1,710,783 – 1,710,858 |
|---|---|
| Length | 75 |
| Sequences | 5 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.44658 |
| G+C content | 0.60368 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -13.28 |
| Energy contribution | -15.04 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 1710783 75 - 22422827 CCACACAGAUACCCGGUAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCUGUUUGUC .......((.((((((...(((....((((((.....)))))).))).)))))).))..(((((....))))).. ( -26.70, z-score = -1.54, R) >droPer1.super_11 880756 68 - 2846995 CCUCACAGAUGG-------CUAAGGACGACGUGCACAGUCCACAGACUUCACGUCCUCUGAGGCUUCGGUGAGUC .(((((.((.((-------((..(((.((((((...((((....)))).)))))).)))..)))))).))))).. ( -27.80, z-score = -2.51, R) >droEre2.scaffold_4644 1691576 71 - 2521924 CCACACAGGUAGCCGGUAACCGAGCAGGAGCUGCACAAGCUCCAUGGCUUGGGUCUCC----ACUCCUGUUUGUC .((.(((((.((..((...((((((.((((((.....))))))...))))))....))----.))))))).)).. ( -31.40, z-score = -3.16, R) >droSec1.super_10 1505631 75 - 3036183 CCACACAGAUAGCCGGCAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCUGUUUGUC ....((((((((((((...))).)).((((((.....))))))(..((.((((....)))).))..)))))))). ( -29.20, z-score = -1.79, R) >droSim1.chrX 1203918 75 - 17042790 CCACACAGAUAGCCGGCAACCGAGCAUGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCUGUUUGUC ....((((((((((((...))).))..(((((.....))))).(..((.((((....)))).))..)))))))). ( -24.80, z-score = -0.90, R) >consensus CCACACAGAUAGCCGGCAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCUGUUUGUC ....................((((((((((((.....((.((((.....)))).)).....)))))))))))).. (-13.28 = -15.04 + 1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:07:31 2011